Comparing PyProBE Performance#
This example will demonstrate the performance benefits of PyProBE against Pandas, a popular library for dataframes.
[1]:
%%capture
%pip install matplotlib
%pip install pandas
[2]:
import os
import timeit
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import polars as pl
import pyprobe
%matplotlib inline
Setting up data analysis in PyProBE requires conversion into the PyProBE format. This is normally the most time-intensive process, but only needs to be performed once.
[3]:
info_dictionary = {
"Name": "Sample cell",
"Chemistry": "NMC622",
"Nominal Capacity [Ah]": 0.04,
"Cycler number": 1,
"Channel number": 1,
}
cell = pyprobe.Cell(info=info_dictionary)
data_directory = "../../../tests/sample_data/neware"
# cell.process_cycler_file(cycler='neware',
# folder_path=data_directory,
# input_filename='sample_data_neware.xlsx',
# output_filename='sample_data_neware.parquet')
We will measure the time for PyProBE and Pandas to read from a parquet file and filter the data a few times. With PyProBE we can call the built-in filtering methods, whereas Pandas must perform the filtering manually.
[4]:
def measure_pyprobe(repeats, file):
steps = 5
cumulative_time = np.zeros((steps, repeats))
for repeat in range(repeats):
start_time = timeit.default_timer()
cell.procedure = {}
cell.add_procedure(
procedure_name="Sample", folder_path=data_directory, filename=file
)
cumulative_time[0, repeat] = timeit.default_timer() - start_time
experiment = cell.procedure["Sample"].experiment("Break-in Cycles")
cumulative_time[1, repeat] = timeit.default_timer() - start_time
cycle = experiment.cycle(1)
cumulative_time[2, repeat] = timeit.default_timer() - start_time
step = cycle.discharge(0)
cumulative_time[3, repeat] = timeit.default_timer() - start_time
time, voltage = step.get("Time [s]", "Voltage [V]")
cumulative_time[4, repeat] = timeit.default_timer() - start_time
return cumulative_time, time, voltage
def measure_pandas(repeats, file, test_csv=True):
steps = 5
cumulative_time = np.zeros((steps, repeats))
csv_time = np.zeros(repeats)
for repeat in range(repeats):
if test_csv:
start_time = timeit.default_timer()
df = pd.read_csv(
data_directory + "/" + str.replace(file, ".parquet", ".csv")
)
csv_time[repeat] = timeit.default_timer() - start_time
start_time = timeit.default_timer()
df = pd.read_parquet(data_directory + "/" + file)
# Add a column to identify the cycle number
df["Cycle"] = (
(df["Step"].astype(int) - df["Step"].astype(int).shift() < 0)
.fillna(0)
.cumsum()
)
cumulative_time[0, repeat] = timeit.default_timer() - start_time
experiment = df[df["Step"].isin([4, 5, 6, 7])]
cumulative_time[1, repeat] = timeit.default_timer() - start_time
unique_cycles = experiment["Cycle"].unique()
cycle = experiment[experiment["Cycle"] == unique_cycles[1]]
cumulative_time[2, repeat] = timeit.default_timer() - start_time
step = cycle[cycle["Current [A]"] < 0]
unique_events = step["Event"].unique()
step = step[step["Event"] == unique_events[0]]
cumulative_time[3, repeat] = timeit.default_timer() - start_time
voltage = step["Voltage [V]"].values
time = step["Time [s]"].values
cumulative_time[4, repeat] = timeit.default_timer() - start_time
return cumulative_time, csv_time, time, voltage
def make_boxplots(total_time_pyprobe, total_time_pandas, log_scale=False):
# Create labels for the boxplots
labels = [
"1: Read file",
"2: Select experiment",
"3: Select cycle",
"4: Select step",
"5: Return voltage",
]
# Create the subplots
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(14, 6), sharey=True)
# Boxplot for pyprobe
ax1.boxplot(
total_time_pyprobe.T,
tick_labels=labels,
vert=True,
patch_artist=True,
showfliers=False,
)
ax1.set_title("PyProBE Execution Time")
ax1.set_ylabel("Cumulative Time (seconds)")
if log_scale:
ax1.set_yscale("log")
# Boxplot for Pandas
ax2.boxplot(
total_time_pandas.T,
tick_labels=labels,
vert=True,
patch_artist=True,
showfliers=False,
)
ax2.set_title("Pandas Execution Time")
ax2.yaxis.set_visible(False) # Remove y-axis on the right-hand subplot
# Adjust layout
plt.tight_layout()
plt.show()
Running the tests shows the initial overhead for PyProBE to read and filter the data is zero. This is because of the Lazy implementation where all the computation is delayed until the final request for data is made. Overall, it is faster than Pandas as the pyprobe backend is able to optimize the filtering process, instead of requiring filters to be performed one-by-one.
[5]:
repeats = 100
total_time_pyprobe, pyprobe_time, pyprobe_voltage = measure_pyprobe(
repeats, "sample_data_neware.parquet"
)
total_time_pandas, csv_time_pandas, pandas_time, pandas_voltage = measure_pandas(
repeats, "sample_data_neware.parquet"
)
make_boxplots(total_time_pyprobe, total_time_pandas)
median_total_time_idx_pyprobe = np.argsort(total_time_pyprobe[-1, :])[
total_time_pyprobe.shape[1] // 2
]
median_total_time_idx_pandas = np.argsort(total_time_pandas[-1, :])[
total_time_pandas.shape[1] // 2
]
median_total_time_pyprobe = total_time_pyprobe[:, median_total_time_idx_pyprobe]
median_total_time_pandas = total_time_pandas[:, median_total_time_idx_pandas]
print(
f"The median execution time for the filtering query in PyProBE is {median_total_time_pandas[-1]/median_total_time_pyprobe[-1]:.2f} times faster than Pandas."
)
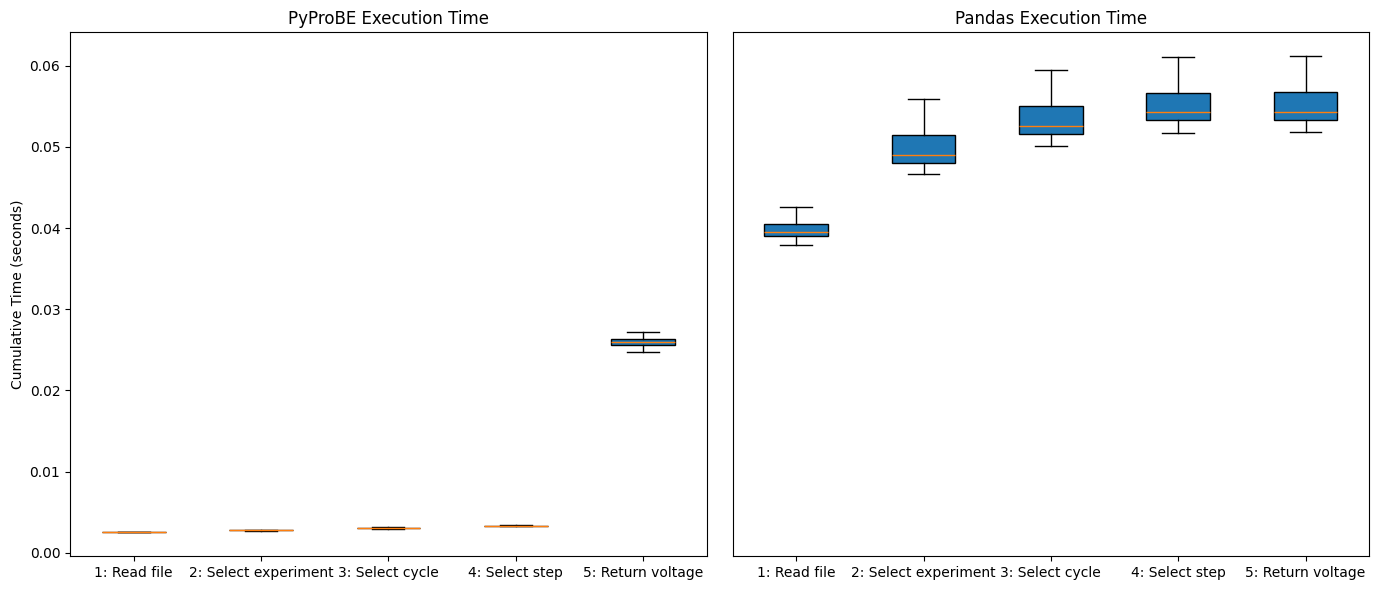
The median execution time for the filtering query in PyProBE is 2.09 times faster than Pandas.
Confirm that the same data has been retrieved:
[6]:
assert np.allclose(pyprobe_voltage, pandas_voltage)
plt.figure(figsize=(10, 6))
plt.plot(pyprobe_time, pyprobe_voltage, label="PyProBE")
plt.plot(pandas_time, pandas_voltage, label="Pandas")
plt.xlabel("Time [s]")
plt.ylabel("Voltage [V]")
plt.legend()
[6]:
<matplotlib.legend.Legend at 0x7f6800b52150>
Much of the performance behind PyProBE is careful selection of the data file format. PyProBE uses the .parquet file format due to its exceptional speed. The results above show pandas reading from the parquet format. The below plot illustrates its performance benefit:
[7]:
plt.figure()
plt.boxplot(
[csv_time_pandas, total_time_pyprobe[-1, :]],
tick_labels=["Read from .csv", "Read from .parquet"],
vert=True,
patch_artist=True,
showfliers=False,
)
plt.ylabel("Time (seconds)")
plt.show()
average_difference = np.median(csv_time_pandas) / np.median(total_time_pyprobe[-1, :])
print(
f"Reading from .parquet is on average {average_difference:.2f} times faster than reading from .csv"
)
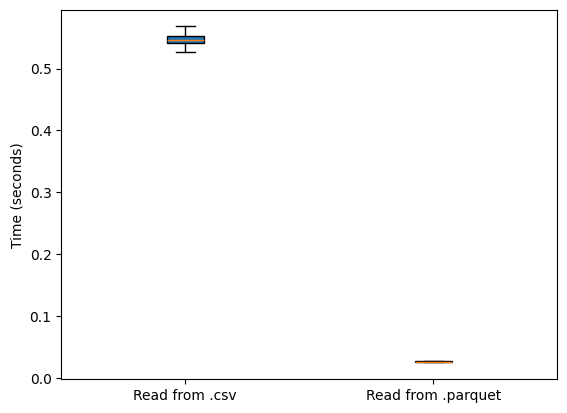
Reading from .parquet is on average 20.99 times faster than reading from .csv
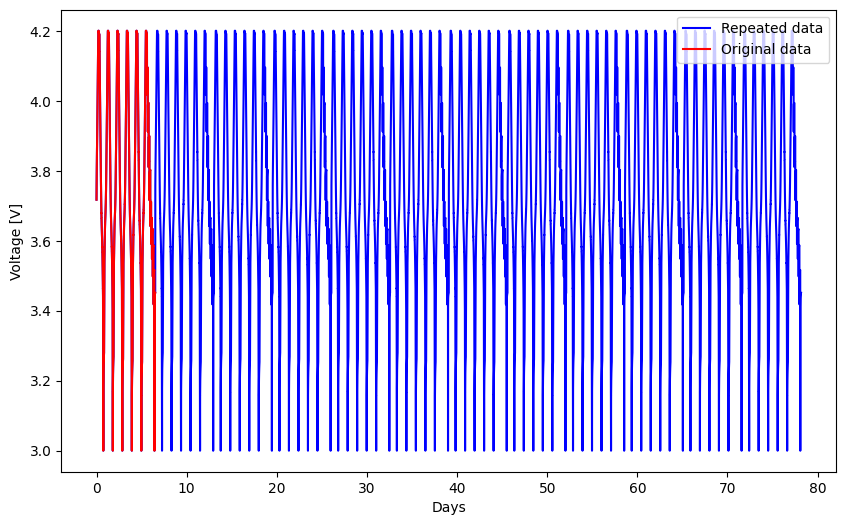
The sample dataset is not large, covering only around a week of testing. Battery degradation experiments can last for years, so the test below demonstrates the scalability of PyProBE to large data sets. We will now repeat our battery experiment multiple times and save the new dataset:
[8]:
# Read the original data from the Parquet file
data = pl.read_parquet(data_directory + "/sample_data_neware.parquet")
# Repeat the data
n_repeats = 12
repeated_data = pl.concat([data] * n_repeats)
# Repeat the 'Cycle' and 'Event' columns to match the length of the repeated data
event_repeated = pl.concat([data["Event"]] * n_repeats)
step_repeated = pl.concat([data["Step"]] * n_repeats)
time_repeated = pl.concat([data["Time [s]"]] * n_repeats)
# Increment the 'Cycle' and 'Event' columns
event_increment = data["Event"].max() + 1
step_increment = data["Step"].max() + 1
time_increment = data["Time [s]"].max()
repeated_data = repeated_data.with_columns(
[
# (pl.arange(0, len(repeated_data)) // len(data) * cycle_increment + cycle_repeated).alias('Cycle'),
(
pl.arange(0, len(repeated_data)) // len(data) * event_increment
+ event_repeated
).alias("Event"),
(
pl.arange(0, len(repeated_data)) // len(data) * event_increment
+ step_repeated
).alias("Step"),
(
pl.arange(0, len(repeated_data)) // len(data) * time_increment
+ time_repeated
).alias("Time [s]"),
]
)
# Write the repeated data to a new Parquet file
repeated_data.write_parquet(data_directory + "/sample_data_neware_repeated.parquet")
# plot the repeated data
plt.figure(figsize=(10, 6))
plt.plot(
repeated_data["Time [s]"] / 3600 / 24,
repeated_data["Voltage [V]"],
label="Repeated data",
color="blue",
)
plt.plot(
data["Time [s]"] / 3600 / 24,
data["Voltage [V]"],
label="Original data",
color="red",
)
plt.xlabel("Days")
plt.ylabel("Voltage [V]")
plt.legend(loc="upper right")
[8]:
<matplotlib.legend.Legend at 0x7f681b438bc0>
And re-run the test:
[9]:
repeats = 100
rep_total_time_pyprobe, rep_pyprobe_time, rep_pyprobe_voltage = measure_pyprobe(
repeats, "sample_data_neware_repeated.parquet"
)
rep_total_time_pandas, rep_csv_time_pandas, rep_pandas_time, rep_pandas_voltage = (
measure_pandas(repeats, "sample_data_neware_repeated.parquet", test_csv=False)
)
rep_median_total_time_idx_pyprobe = np.argsort(rep_total_time_pyprobe[-1, :])[
rep_total_time_pyprobe.shape[1] // 2
]
rep_median_total_time_idx_pandas = np.argsort(rep_total_time_pandas[-1, :])[
rep_total_time_pandas.shape[1] // 2
]
rep_median_total_time_pyprobe = rep_total_time_pyprobe[
:, rep_median_total_time_idx_pyprobe
]
rep_median_total_time_pandas = rep_total_time_pandas[
:, rep_median_total_time_idx_pandas
]
os.remove(data_directory + "/sample_data_neware_repeated.parquet")
print(
f"The median execution time for the filtering query in PyProBE is {rep_median_total_time_pandas[-1]/rep_median_total_time_pyprobe[-1]:.2f} times faster than Pandas."
)
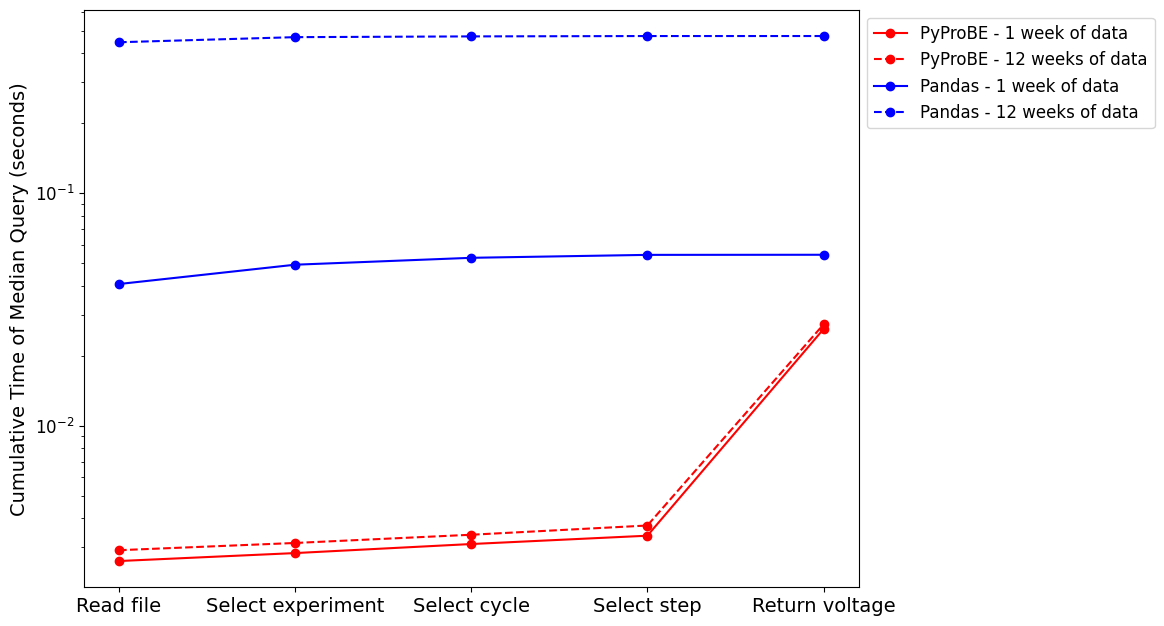
plt.figure(figsize=(10, 7.5))
plt.plot(
median_total_time_pyprobe, label="PyProBE - 1 week of data", marker="o", color="red"
)
plt.plot(
rep_median_total_time_pyprobe,
label=f"PyProBE - {n_repeats} weeks of data",
marker="o",
linestyle="--",
color="red",
)
plt.plot(
median_total_time_pandas, label="Pandas - 1 week of data", marker="o", color="blue"
)
plt.plot(
rep_median_total_time_pandas,
label=f"Pandas - {n_repeats} weeks of data",
marker="o",
linestyle="--",
color="blue",
)
plt.yscale("log")
plt.xticks(
range(5),
["Read file", "Select experiment", "Select cycle", "Select step", "Return voltage"],
fontsize=14,
)
plt.yticks(fontsize=12)
plt.legend(bbox_to_anchor=(1, 1), fontsize=12)
plt.ylabel("Cumulative Time of Median Query (seconds)", fontsize=14)
data = {
"Period": ["1 week", f"{n_repeats} weeks", "Increase Factor"],
"PyProBE": [
median_total_time_pyprobe[-1],
rep_median_total_time_pyprobe[-1],
rep_median_total_time_pyprobe[-1] / median_total_time_pyprobe[-1],
],
"Pandas": [
median_total_time_pandas[-1],
rep_median_total_time_pandas[-1],
rep_median_total_time_pandas[-1] / median_total_time_pandas[-1],
],
}
print("Execution time breakdown:")
df = pd.DataFrame(data)
print(df)
The median execution time for the filtering query in PyProBE is 17.42 times faster than Pandas.
Execution time breakdown:
Period PyProBE Pandas
0 1 week 0.026024 0.054350
1 12 weeks 0.027253 0.474633
2 Increase Factor 1.047193 8.732911
Confirm that the same data has been retrieved:
[10]:
assert np.allclose(rep_pyprobe_voltage, rep_pandas_voltage)
plt.figure(figsize=(10, 6))
plt.plot(rep_pyprobe_time, rep_pyprobe_voltage, label="PyProBE")
plt.plot(rep_pandas_time, rep_pandas_voltage, label="Pandas")
plt.xlabel("Time [s]")
plt.ylabel("Voltage [V]")
plt.legend()
[10]:
<matplotlib.legend.Legend at 0x7f68180ccda0>
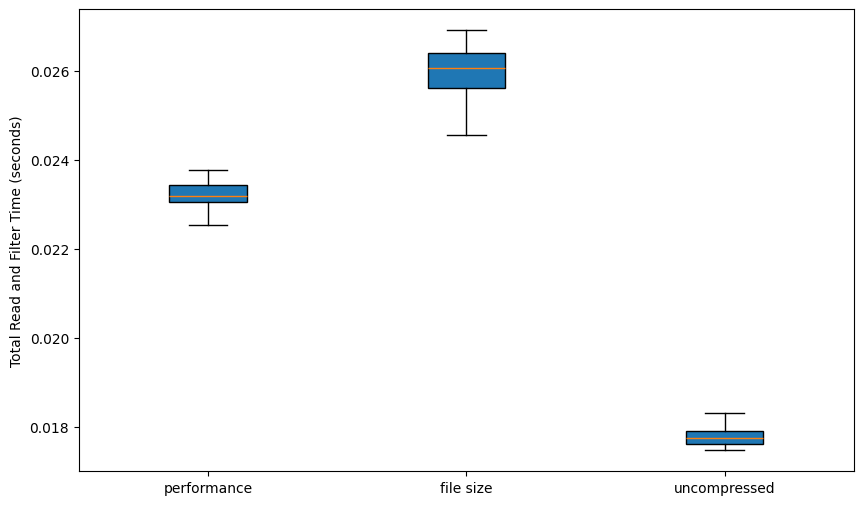
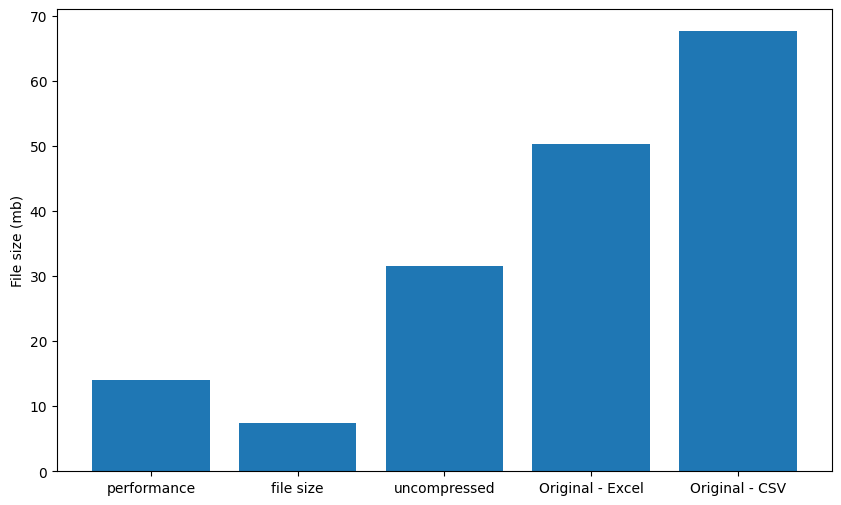
Parquet files have a number of options for compression. In PyProBE, two have been selected as options when processing a cycler file. Along with uncompressed, you can prioritise either file size or performance, with performance being the default. The compression options are benchmarked below:
[11]:
compression_priority = ["performance", "file size", "uncompressed"]
times = np.zeros((len(compression_priority), repeats))
file_sizes = np.zeros((len(compression_priority)))
for i, priority in enumerate(compression_priority):
cell.process_cycler_file(
cycler="neware",
folder_path=data_directory,
input_filename="sample_data_neware.xlsx",
output_filename="sample_data_neware_test.parquet",
overwrite_existing=True,
compression_priority=priority,
)
file_sizes[i] = os.path.getsize(
data_directory + f"/sample_data_neware_test.parquet"
)
pyprobe_time, _, _ = measure_pyprobe(repeats, "sample_data_neware_test.parquet")
times[i, :] = pyprobe_time[-1, :]
file_sizes = np.append(
file_sizes,
[
os.path.getsize(data_directory + f"/sample_data_neware.xlsx"),
os.path.getsize(data_directory + f"/sample_data_neware.csv"),
],
)
plt.figure(figsize=(10, 6))
plt.boxplot(
times.T,
tick_labels=compression_priority,
vert=True,
patch_artist=True,
showfliers=False,
)
plt.ylabel("Total Read and Filter Time (seconds)")
plt.show()
plt.figure(figsize=(10, 6))
plt.bar(
list(compression_priority + ["Original - Excel", "Original - CSV"]),
file_sizes / 10**6,
)
plt.ylabel("File size (mb)")
plt.show()
os.remove(data_directory + "/sample_data_neware_test.parquet")