A brief introduction to RNA-sequencing
RNA is a nucleic acid that is similar to DNA. In cells, DNA acts as a long-term storage of genetic information. RNA, on the other hand, has very different functions. One form of RNA, messenger RNA (mRNA), copies the genetic information encoded as DNA and carries it out of the nucleus; here, the cell can use the mRNA sequences to synthesise proteins. When the information from a gene is copied by RNA, we say that the gene has been expressed. This flow of genetic information from DNA to RNA to protein is known as the central dogma of molecular biology.
With the advent of the Human Genome Project, we can now determine the sequence of an organism's entire genome. This information has been invaluable to life science researchers; for instance, genome-wide association studies are used to identify genetic mutations that are associated with traits, such as diseases. However, the genome remains relatively static throughout the life of most organisms, so it cannot tell us about the current state of a biological system. The expression of genes, on the other hand, is a lot more dynamic and will change in response to stimuli. For instance, we could identify genes that are activated in a particular disease by comparing the gene expression in healthy people to individuals with the disease.
RNA sequencing allows us to elucidate the sequence of a set of RNA molecules in a sample. Given that we know the genome sequence of the organism we are studying, we can find out where each RNA molecule came from in the genome. We are commonly most interested in mRNA molecules, which copy the sequences of genes and use this information to synthesise proteins in the cell. By counting the number of sequences that map back to each gene in an organism, we can quantify gene expression for the genes. Having done these processing stages, we can use various downstream analysis strategies to learn more about the biology of a system. For more information, you can explore the notebook docs/downstream_analysis.md in this repository.
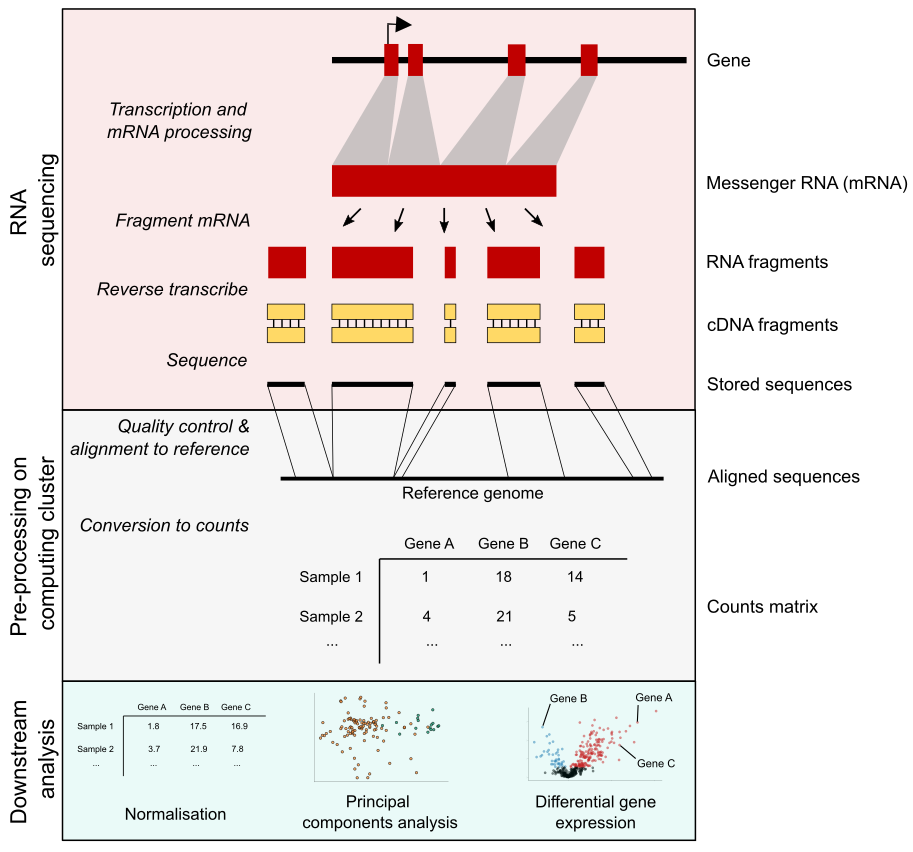
The diagram below summarises the process of an RNA-seq experiment. We consider there to be three main stages:
- Performing RNA sequencing. In this stage we generate a list of sequences in a sample.
- Processing the data. This process could take many forms, but we will focus on quantifying the number of RNA sequences that originated from each gene. This notebook will focus on this stage, using simple bash scripts and open source tools to perform the data processing steps.
- Analysing the data. There are lots of ways we could analyse the data, but we will focus on using R to perform some of the most basic and popular analyses. See the
docs/downstream_analysis.mdnotebook for more details.
If you are interested in learning more about RNA-seq and other methods for measuring gene expression, you could start with the following review. This video also provides a good introduction to the technique.