What is nextflow?
nextflow is a workflow management system that allows users to orchestrate the running of scripts into a single cohesive pipeline. nextflow pipelines allow reproducibility and can be easily shared and used by others. nextflow is a DSL (domain specific language) with simple methods for combining scripts into fully-fledged pipelines. For detailed information, see the nextflow site.
In this document we will discuss features of nextflow that are useful for coordinating our simple RNA-seq pipeline, but these explanations will be brief and will only scratch the surface of what nextflow can do. We will explain the main concepts alongside the building of our simple RNA-seq pipeline. If you want a more in-depth explanation of writing nextflow code, you can read the nextflow documentation or look for more detailed guides.
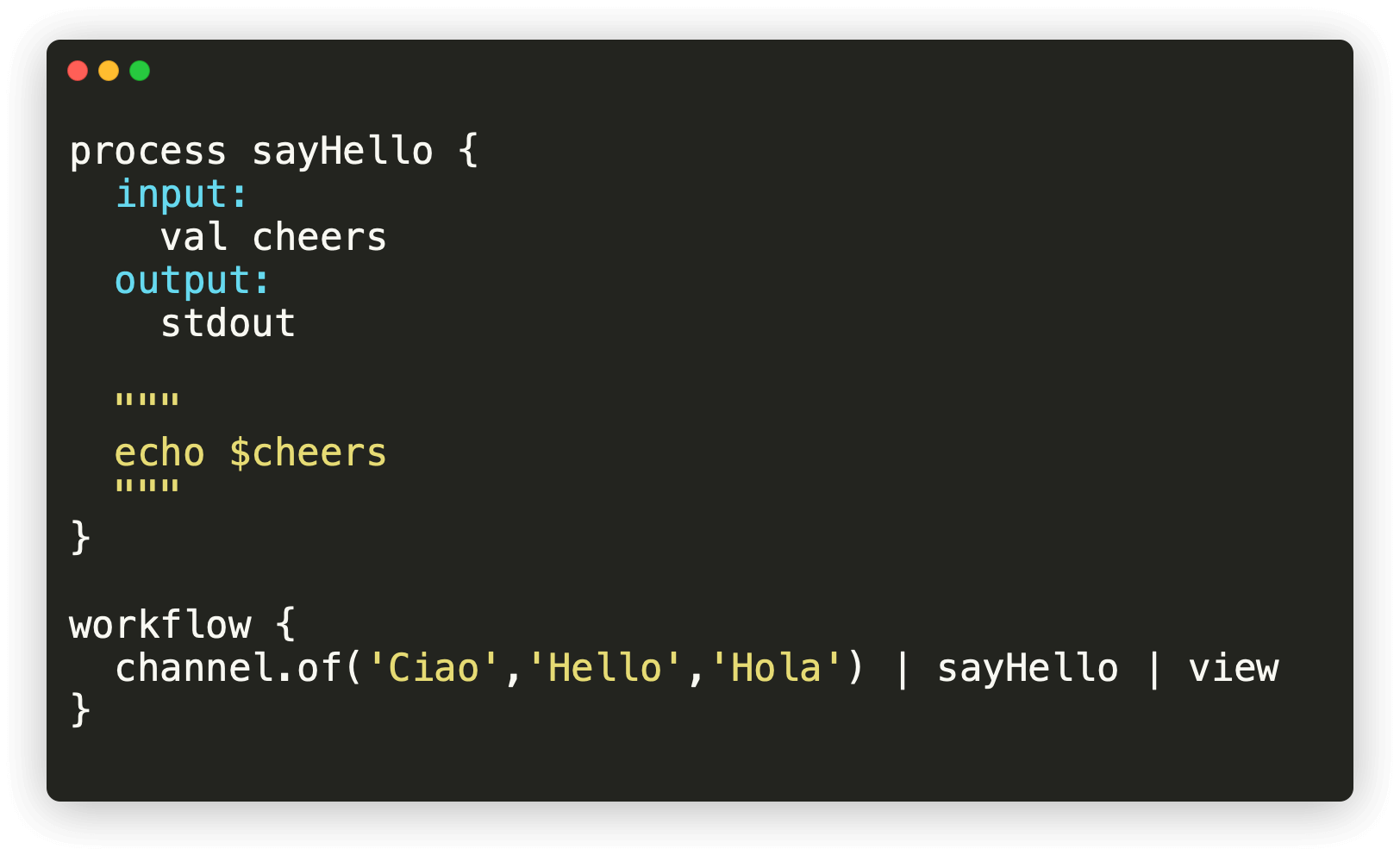
nextflow can integrate lots of different types of scripts, including those written in bash, Python and R. So, you aren't required to fully learn a new language in order to create nextflow pipelines, you can instead reuse existing scripts and workflows. Our previous pipelines both made use of the same scripts in the bin/ directory, but they used them in slightly different ways. In this document we will use these same scripts, but we will orchestrate them using nextflow.
When we wrote the simple local pipeline and the parallelised pipeline, we had to modify the scripts in order to make them compatible with the Imperial cluster. If we were to send this pipeline to a researcher at another university, they might have some difficulty adapting it. nextflow is aware of common cluster schedulers, like PBS and slurm, so it can worry about queueing jobs on the cluster for us! We can write code that runs on our local computer, then export it to a computing cluster to do the heavy lifting without changing anything about the pipeline itself. This is possible because we can give nextflow a config file specifying how it can interact with our specific computing cluster. Many institutions have pre-built config files, so you may not even need to write your own!
For our pipeline, we are largely motivated to use nextflow because: i) we don't want to have to worry about setting up our scripts as jobs on the cluster, like we did in docs/parallelised_pipeline.md; ii) setting up our pipeline with nextflow will make it easier for others to run it, especially if they use a different computing cluster; iii) nextflow will make sure each processing stage produces the expected outputs, and make it far easier to identify the source of the problem.
We will also see that nextflow makes it easier to work with environment managers like conda.