Basic output plots
Nathan Green
2018-04-04
Introduction
We combine the cost and health results from the screening, testing and treatment decision tree with the cost and health results from the population model follow-up to defined end-point. We then plot some basic cost-effectiveness planes.
Load packages
library(treeSimR)
library(LTBIscreeningproject)
library(dplyr)
library(data.tree)
library(purrr)
library(tibble)
library(magrittr)
library(plotCostEffectiveness)
library(ggplot2)
library(BCEA)Load in data
data("scenario_parameters", package = "LTBIscreeningproject")
load("aTB_CE_stats.RData")
dectree_res <- readRDS("decision_tree_output.Rds")
cohort <- readRDS("cohort.Rds")Create input matrices
We’re going to use the BCEA package to calculate some additional values and do some of the plotting. In order to use this we need to rearrange some of our data.
# discount due to delay to screening
##TODO: what is actual number?
screen_discount <- 0.9
scenario.names <-
c(0, seq_len(length(dectree_res))) %>%
as.character(.)Cost and QALY gain due to active TB in the population
tb_cost <-
aTB_CE_stats$cost_incur_person %>%
do.call(cbind.data.frame, .) %>%
add_column('0' = 0, .before = 1) %>%
set_names(nm = scenario.names)
tb_QALYgain <-
aTB_CE_stats$QALYgain_person %>%
do.call(cbind.data.frame, .) %>%
add_column('0' = 0, .before = 1) %>%
set_names(nm = scenario.names)Cost and QALY gain due to screening
LTBI_cost <-
purrr::map(dectree_res, "mc_cost") %>%
do.call(cbind.data.frame, .) %>%
multiply_by(screen_discount) %>%
add_column('0' = 0, .before = 1)
LTBI_QALYgain <-
purrr::map(dectree_res, "mc_health") %>%
do.call(cbind.data.frame, .) %>%
multiply_by(-screen_discount) %>%
add_column('0' = 0, .before = 1)
c.total <- as.matrix(LTBI_cost + tb_cost)
e.total <- as.matrix(LTBI_QALYgain + tb_QALYgain)
c.total
#> 0 1 2 3
#> [1,] 0 19.44459 26.47664 35.55441
#> [2,] 0 24.96156 27.25336 23.87494
#> [3,] 0 24.67908 15.81333 31.24292
#> [4,] 0 28.32432 10.58710 32.19658
#> [5,] 0 22.88430 34.47200 31.72002
#> [6,] 0 29.46146 29.79122 12.53047
#> [7,] 0 14.11653 17.76350 49.51426
#> [8,] 0 19.40375 38.81264 37.04938
#> [9,] 0 24.86463 39.49851 33.42976
#> [10,] 0 21.29811 20.15852 37.67071
e.total
#> 0 1 2 3
#> [1,] 0 0.0009093726 8.824310e-04 9.067279e-04
#> [2,] 0 0.0006215663 8.837187e-04 1.220403e-03
#> [3,] 0 0.0006222185 9.087648e-04 9.240392e-04
#> [4,] 0 0.0006143797 1.840528e-03 1.219264e-03
#> [5,] 0 0.0005963736 2.814660e-04 5.859530e-04
#> [6,] 0 0.0005960537 8.826764e-04 2.448010e-03
#> [7,] 0 0.0012477170 1.534320e-03 -7.919966e-06
#> [8,] 0 0.0009092369 3.063636e-04 1.558076e-03
#> [9,] 0 0.0009342820 -6.393039e-06 1.220447e-03
#> [10,] 0 0.0006216804 1.221663e-03 5.932782e-04Cost-effectiveness planes
screen.bcea <- BCEA::bcea(e = -e.total, # Q1 - Q0 different way round in function!
c = -c.total,
ref = 1,
interventions = colnames(e.total))
cbPalette <- colorRampPalette(c("red", "orange", "green", "blue"))(screen.bcea$n.comparisons)
gg <- contour2(screen.bcea, graph = "ggplot2", wtp = 20000)
gg + scale_colour_manual(values = cbPalette)
#> Scale for 'colour' is already present. Adding another scale for
#> 'colour', which will replace the existing scale.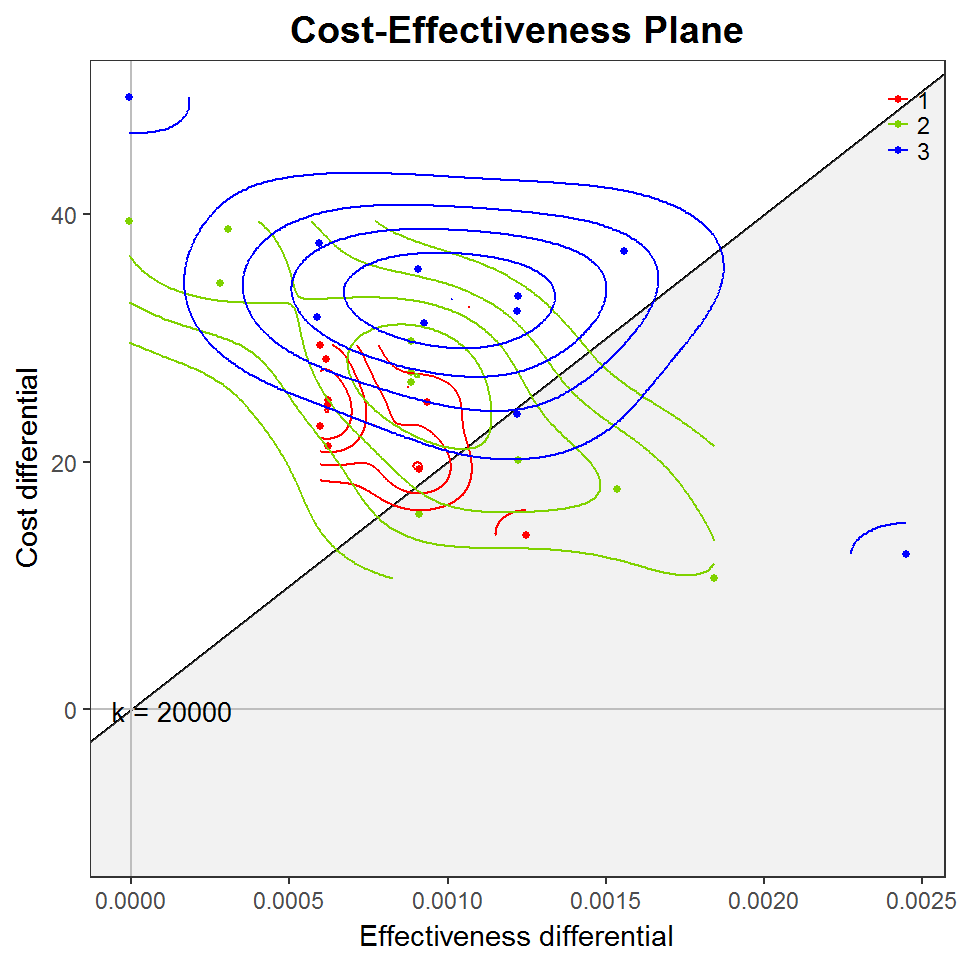
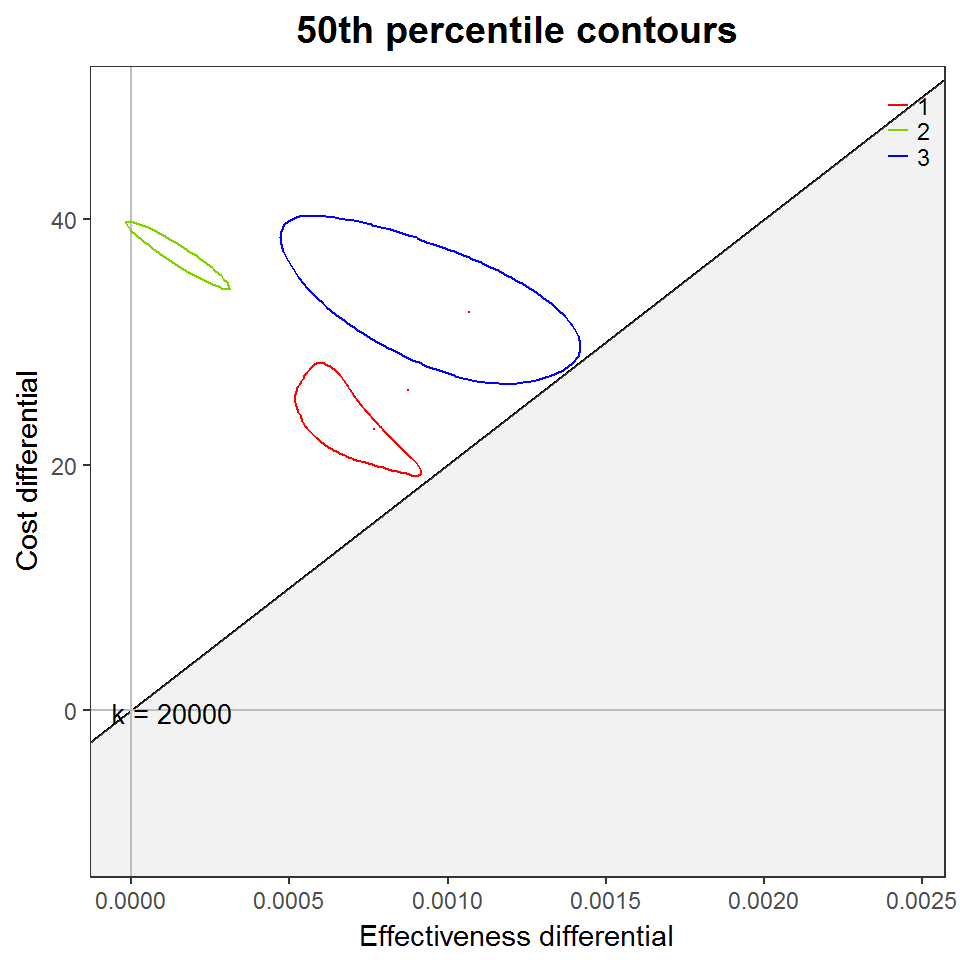
my_contour2(screen.bcea,
graph = "ggplot2",
wtp = 20000,
CONTOUR_PC = "50%") +
ggtitle('50th percentile contours') +
scale_colour_manual(values = cbPalette)
#> Scale for 'colour' is already present. Adding another scale for
#> 'colour', which will replace the existing scale.