 |
ICFERST
25-11
Reservoir simulator based on DCVFEM, Dynamic Mesh optimisation and Surface-based modelling
|
 |
ICFERST
25-11
Reservoir simulator based on DCVFEM, Dynamic Mesh optimisation and Surface-based modelling
|
Fortran Documentation for the geochemical reaction module PhreeqcRM. More...
Functions/Subroutines | |
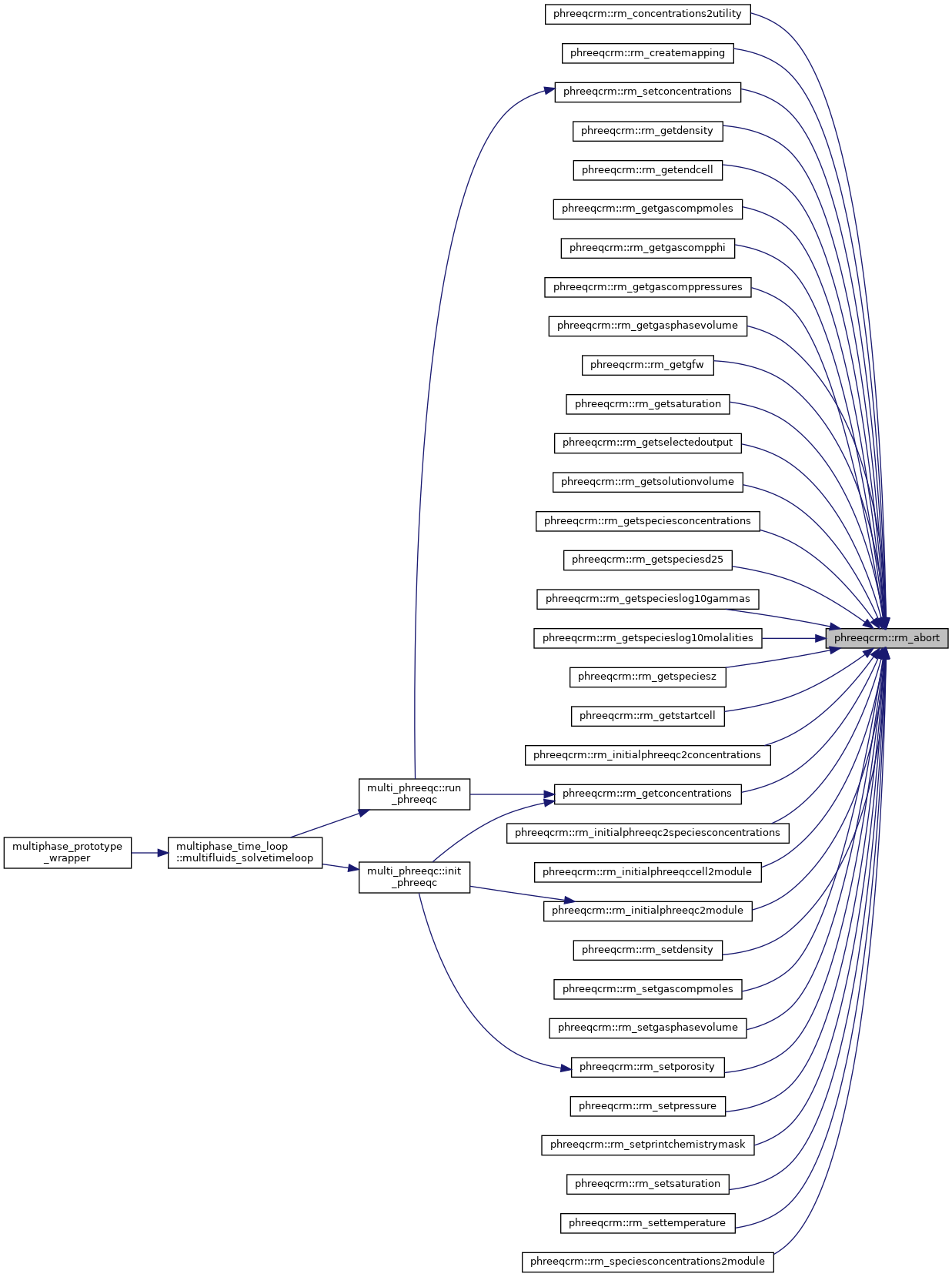
| integer function | rm_abort (id, irm_result, err_str) |
| Abort the program. irm_result will be interpreted as an IRM_RESULT value and decoded; err_str will be printed; and the reaction module will be destroyed. If using MPI, an MPI_Abort message will be sent before the reaction module is destroyed. If the id is an invalid instance, RM_Abort will return a value of IRM_BADINSTANCE, otherwise the program will exit with a return code of 4. More... | |
| integer function | rm_closefiles (id) |
| Close the output and log files. More... | |
| integer function | rm_concentrations2utility (id, c, n, tc, p_atm) |
| N sets of component concentrations are converted to SOLUTIONs numbered 1-n in the Utility IPhreeqc. The solutions can be reacted and manipulated with the methods of IPhreeqc. If solution concentration units (RM_SetUnitsSolution) are per liter, one liter of solution is created in the Utility instance; if solution concentration units are mass fraction, one kilogram of solution is created in the Utility instance. The motivation for this method is the mixing of solutions in wells, where it may be necessary to calculate solution properties (pH for example) or react the mixture to form scale minerals. The code fragments below make a mixture of concentrations and then calculate the pH of the mixture. More... | |
| integer function | rm_create (nxyz, nthreads) |
| Creates a reaction module. If the code is compiled with the preprocessor directive USE_OPENMP, the reaction module is multithreaded. If the code is compiled with the preprocessor directive USE_MPI, the reaction module will use MPI and multiple processes. If neither preprocessor directive is used, the reaction module will be serial (unparallelized). More... | |
| integer function | rm_createmapping (id, grid2chem) |
| Provides a mapping from grid cells in the user's model to reaction cells in PhreeqcRM. The mapping is used to eliminate inactive cells and to use symmetry to decrease the number of cells for which chemistry must be run. The array grid2chem of size nxyz (the number of grid cells, RM_GetGridCellCount) must contain the set of all integers 0 <= i < count_chemistry, where count_chemistry is a number less than or equal to nxyz. Inactive cells are assigned a negative integer. The mapping may be many-to-one to account for symmetry. Default is a one-to-one mapping–all user grid cells are reaction cells (equivalent to grid2chem values of 0,1,2,3,...,nxyz-1). More... | |
| integer function | rm_decodeerror (id, e) |
| If e is negative, this method prints an error message corresponding to IRM_RESULT e. If e is non-negative, no action is taken. More... | |
| integer function | rm_destroy (id) |
| Destroys a reaction module. More... | |
| integer function | rm_dumpmodule (id, dump_on, append) |
| Writes the contents of all workers to file in _RAW formats, including SOLUTIONs and all reactants. More... | |
| integer function | rm_errormessage (id, errstr) |
| Send an error message to the screen, the output file, and the log file. More... | |
| integer function | rm_findcomponents (id) |
| Returns the number of items in the list of all elements in the InitialPhreeqc instance. Elements are those that have been defined in a solution or any other reactant (EQUILIBRIUM_PHASE, KINETICS, and others). The method can be called multiple times and the list that is created is cummulative. The list is the set of components that needs to be transported. By default the list includes water, excess H and excess O (the H and O not contained in water); alternatively, the list may be set to contain total H and total O (RM_SetComponentH2O), which requires transport results to be accurate to eight or nine significant digits. If multicomponent diffusion (MCD) is to be modeled, there is a capability to retrieve aqueous species concentrations (RM_GetSpeciesConcentrations) and to set new solution concentrations after MCD by using individual species concentrations (RM_SpeciesConcentrations2Module). To use these methods the save-species property needs to be turned on (RM_SetSpeciesSaveOn). If the save-species property is on, RM_FindComponents will generate a list of aqueous species (RM_GetSpeciesCount, RM_GetSpeciesName), their diffusion coefficients at 25 C (RM_GetSpeciesD25), their charge (RM_GetSpeciesZ). More... | |
| integer function | rm_getbackwardmapping (id, n, list, size) |
| Fills an array with the cell numbers in the user's numbering sytstem that map to a cell in the PhreeqcRM numbering system. The mapping is defined by RM_CreateMapping. More... | |
| integer function | rm_getchemistrycellcount (id) |
| Returns the number of chemistry cells in the reaction module. The number of chemistry cells is defined by the set of non-negative integers in the mapping from user grid cells (RM_CreateMapping). The number of chemistry cells is less than or equal to the number of cells in the user's model. More... | |
| integer function | rm_getcomponent (id, num, comp_name) |
| Retrieves an item from the reaction-module component list that was generated by calls to RM_FindComponents. More... | |
| integer function | rm_getcomponentcount (id) |
| Returns the number of components in the reaction-module component list. The component list is generated by calls to RM_FindComponents. The return value from the last call to RM_FindComponents is equal to the return value from RM_GetComponentCount. More... | |
| integer function | rm_getconcentrations (id, c) |
| Transfer solution concentrations from each reaction cell to the concentration array given in the argument list (c). Units of concentration for c are defined by RM_SetUnitsSolution. For concentration units of per liter, the solution volume is used to calculate the concentrations for c. For mass fraction concentration units, the solution mass is used to calculate concentrations for c. Two options are available for the volume and mass of solution that are used in converting to transport concentrations: (1) the volume and mass of solution are calculated by PHREEQC, or (2) the volume of solution is the product of saturation (RM_SetSaturation), porosity (RM_SetPorosity), and representative volume (RM_SetRepresentativeVolume), and the mass of solution is volume times density as defined by RM_SetDensity. RM_UseSolutionDensityVolume determines which option is used. For option 1, the databases that have partial molar volume definitions needed to accurately calculate solution volume are phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_getdensity (id, density) |
| Transfer solution densities from the reaction cells to the array given in the argument list (density). Densities are those calculated by the reaction module. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate density: phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_getendcell (id, ec) |
| Returns an array with the ending cell numbers from the range of cell numbers assigned to each worker. More... | |
| integer function | rm_getequilibriumphasescount (id) |
| Returns the number of equilibrium phases in the initial-phreeqc module. RM_FindComponents must be called before RM_GetEquilibriumPhasesCount. This method may be useful when generating selected output definitions related to equilibrium phases. More... | |
| integer function | rm_getequilibriumphasesname (id, num, name) |
| Retrieves an item from the equilibrium phase list. The list includes all phases included in any EQUILIBRIUM_PHASES definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetEquilibriumPhasesName. This method may be useful when generating selected output definitions related to equilibrium phases. More... | |
| integer function | rm_geterrorstring (id, errstr) |
| Returns a string containing error messages related to the last call to a PhreeqcRM method to the character argument (errstr). More... | |
| integer function | rm_geterrorstringlength (id) |
| Returns the length of the string that contains error messages related to the last call to a PhreeqcRM method. More... | |
| integer function | rm_getexchangename (id, num, name) |
| Retrieves an item from the exchange name list. RM_FindComponents must be called before RM_GetExchangeName. The exchange names vector is the same length as the exchange species names vector and provides the corresponding exchange site (for example, X corresponing to NaX). This method may be useful when generating selected output definitions related to exchangers. More... | |
| integer function | rm_getexchangespeciescount (id) |
| Returns the number of exchange species in the initial-phreeqc module. RM_FindComponents must be called before RM_GetExchangeSpeciesCount. This method may be useful when generating selected output definitions related to exchangers. More... | |
| integer function | rm_getexchangespeciesname (id, num, name) |
| Retrieves an item from the exchange species list. The list of exchange species (such as "NaX") is derived from the list of components (RM_FindComponents) and the list of all exchange names (such as "X") that are included in EXCHANGE definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetExchangeSpeciesName. This method may be useful when generating selected output definitions related to exchangers. More... | |
| integer function | rm_getfileprefix (id, prefix) |
| Returns the reaction-module file prefix to the character argument (prefix). More... | |
| integer function | rm_getgascomponentscount (id) |
| Returns the number of gas phase components in the initial-phreeqc module. RM_FindComponents must be called before RM_GetGasComponentsCount. This method may be useful when generating selected output definitions related to gas phases. More... | |
| integer function | rm_getgascomponentsname (id, num, name) |
| Retrieves an item from the gas components list. The list includes all gas components included in any GAS_PHASE definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetGasComponentsName. This method may be useful when generating selected output definitions related to gas phases. More... | |
| integer function | rm_getgascompmoles (id, gas_moles) |
| Transfer moles of gas components from each reaction cell to the array given in the argument list (gas_moles). More... | |
| integer function | rm_getgascomppressures (id, gas_p) |
| Transfer pressures of gas components from each reaction cell to the array given in the argument list (gas_p). More... | |
| integer function | rm_getgascompphi (id, gas_phi) |
| Transfer fugacity coefficients (phi) of gas components from each reaction cell to the array given in the argument list (gas_phi). Fugacity of a gas component is equal to the pressure of the component times the fugacity coefficient. More... | |
| integer function | rm_getgasphasevolume (id, gas_volume) |
| Transfer volume of gas from each reaction cell to the vector given in the argument list (gas_volume). More... | |
| integer function | rm_getgfw (id, gfw) |
| Returns the gram formula weights (g/mol) for the components in the reaction-module component list. More... | |
| integer function | rm_getgridcellcount (id) |
| Returns the number of grid cells in the user's model, which is defined in the call to RM_Create. The mapping from grid cells to reaction cells is defined by RM_CreateMapping. The number of reaction cells may be less than the number of grid cells if there are inactive regions or symmetry in the model definition. More... | |
| integer function | rm_getiphreeqcid (id, i) |
| Returns an IPhreeqc id for the ith IPhreeqc instance in the reaction module. For the threaded version, there are nthreads + 2 IPhreeqc instances, where nthreads is defined in the constructor (RM_Create). The number of threads can be determined by RM_GetThreadCount. The first nthreads (0 based) instances will be the workers, the next (nthreads) is the InitialPhreeqc instance, and the next (nthreads + 1) is the Utility instance. Getting the IPhreeqc pointer for one of these instances allows the user to use any of the IPhreeqc methods on that instance. For MPI, each process has exactly three IPhreeqc instances, one worker (number 0), one InitialPhreeqc instance (number 1), and one Utility instance (number 2). More... | |
| integer function | rm_getkineticreactionscount (id) |
| Returns the number of kinetic reactions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetKineticReactionsCount. This method may be useful when generating selected output definitions related to kinetic reactions. More... | |
| integer function | rm_getkineticreactionsname (id, num, name) |
| Retrieves an item from the kinetic reactions list. The list includes all kinetic reactions included in any KINETICS definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetKineticReactionsName. This method may be useful when generating selected output definitions related to kinetic reactions. More... | |
| integer function | rm_getmpimyself (id) |
| Returns the MPI task number. For the OPENMP version, the task number is always zero and the result of RM_GetMpiTasks is one. For the MPI version, the root task number is zero, and all workers have a task number greater than zero. The number of tasks can be obtained with RM_GetMpiTasks. The number of tasks and computer hosts are determined at run time by the mpiexec command, and the number of reaction-module processes is defined by the communicator used in constructing the reaction modules (RM_Create). More... | |
| integer function | rm_getmpitasks (id) |
| Returns the number of MPI processes (tasks) assigned to the reaction module. For the OPENMP version, the number of tasks is always one (although there may be multiple threads, RM_GetThreadCount), and the task number returned by RM_GetMpiMyself is zero. For the MPI version, the number of tasks and computer hosts are determined at run time by the mpiexec command. An MPI communicator is used in constructing reaction modules for MPI. The communicator may define a subset of the total number of MPI processes. The root task number is zero, and all workers have a task number greater than zero. More... | |
| integer function | rm_getnthselectedoutputusernumber (id, n) |
| Returns the user number for the nth selected-output definition. Definitions are sorted by user number. Phreeqc allows multiple selected-output definitions, each of which is assigned a nonnegative integer identifier by the user. The number of definitions can be obtained by RM_GetSelectedOutputCount. To cycle through all of the definitions, RM_GetNthSelectedOutputUserNumber can be used to identify the user number for each selected-output definition in sequence. RM_SetCurrentSelectedOutputUserNumber is then used to select that user number for selected-output processing. More... | |
| integer function | rm_getsaturation (id, sat_calc) |
| Returns a vector of saturations (sat_calc) as calculated by the reaction module. Reactions will change the volume of solution in a cell. The transport code must decide whether to ignore or account for this change in solution volume due to reactions. Following reactions, the cell saturation is calculated as solution volume (RM_GetSolutionVolume) divided by the product of representative volume (RM_SetRepresentativeVolume) and the porosity (RM_SetPorosity). The cell saturation returned by RM_GetSaturation may be less than or greater than the saturation set by the transport code (RM_SetSaturation), and may be greater than or less than 1.0, even in fully saturated simulations. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate solution volume and saturation: phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_getselectedoutput (id, so) |
| Populates an array with values from the current selected-output definition. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used to populate the array. More... | |
| integer function | rm_getselectedoutputcolumncount (id) |
| Returns the number of columns in the current selected-output definition. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used. More... | |
| integer function | rm_getselectedoutputcount (id) |
| Returns the number of selected-output definitions. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used. More... | |
| integer function | rm_getselectedoutputheading (id, icol, heading) |
| Returns a selected output heading. The number of headings is determined by RM_GetSelectedOutputColumnCount. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used. More... | |
| integer function | rm_getselectedoutputrowcount (id) |
| Returns the number of rows in the current selected-output definition. However, the method is included only for convenience; the number of rows is always equal to the number of grid cells in the user's model, and is equal to RM_GetGridCellCount. More... | |
| integer function | rm_getsicount (id) |
| Returns the number of phases in the initial-phreeqc module for which saturation indices can be calculated. RM_FindComponents must be called before RM_GetSICount. This method may be useful when generating selected output definitions related to saturation indices. More... | |
| integer function | rm_getsiname (id, num, name) |
| Retrieves an item from the list of all phases for which saturation indices can be calculated. The list includes all phases that contain only elements included in the components in the initial-phreeqc module. The list assumes that all components are present to be able to calculate the entire list of SIs; it may be that one or more components are missing in any specific cell. RM_FindComponents must be called before RM_GetSIName. This method may be useful when generating selected output definitions related to saturation indices. More... | |
| integer function | rm_getsolidsolutioncomponentscount (id) |
| Returns the number of solid solution components in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSolidSolutionComponentsCount. This method may be useful when generating selected output definitions related to solid solutions. More... | |
| integer function | rm_getsolidsolutioncomponentsname (id, num, name) |
| Retrieves an item from the solid solution components list. The list includes all solid solution components included in any SOLID_SOLUTIONS definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSolidSolutionComponentsName. This method may be useful when generating selected output definitions related to solid solutions. More... | |
| integer function | rm_getsolidsolutionname (id, num, name) |
| Retrieves an item from the solid solution names list. The list includes solid solution names included in SOLID_SOLUTIONS definitions in the initial-phreeqc module. The solid solution names vector is the same length as the solid solution components vector and provides the corresponding name of solid solution containing the component. RM_FindComponents must be called before RM_GetSolidSolutionName. This method may be useful when generating selected output definitions related to solid solutions. More... | |
| integer function | rm_getsolutionvolume (id, vol) |
| Transfer solution volumes from the reaction cells to the array given in the argument list (vol). Solution volumes are those calculated by the reaction module. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate solution volume: phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_getspeciesconcentrations (id, species_conc) |
| Transfer concentrations of aqueous species to the array argument (species_conc) This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. Solution volumes used to calculate mol/L are calculated by the reaction module. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate solution volume: phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_getspeciescount (id) |
| The number of aqueous species used in the reaction module. This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. More... | |
| integer function | rm_getspeciesd25 (id, diffc) |
| Transfers diffusion coefficients at 25C to the array argument (diffc). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. Diffusion coefficients are defined in SOLUTION_SPECIES data blocks, normally in the database file. Databases distributed with the reaction module that have diffusion coefficients defined are phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_getspecieslog10gammas (id, species_log10gammas) |
| Transfer log10 aqueous-species activity coefficients to the array argument (species_log10gammas) This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. More... | |
| integer function | rm_getspecieslog10molalities (id, species_log10molalities) |
| Transfer log10 aqueous-species log10 molalities to the array argument (species_log10molalities) To use this method RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. More... | |
| integer function | rm_getspeciesname (id, i, name) |
| Transfers the name of the ith aqueous species to the character argument (name). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. More... | |
| integer function | rm_getspeciessaveon (id) |
| Returns the value of the species-save property. By default, concentrations of aqueous species are not saved. Setting the species-save property to true allows aqueous species concentrations to be retrieved with RM_GetSpeciesConcentrations, and solution compositions to be set with RM_SpeciesConcentrations2Module. More... | |
| integer function | rm_getspeciesz (id, z) |
| Transfers the charge of each aqueous species to the array argument (z). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. More... | |
| integer function | rm_getstartcell (id, sc) |
| Returns an array with the starting cell numbers from the range of cell numbers assigned to each worker. More... | |
| integer function | rm_getsurfacename (id, num, name) |
| Retrieves the surface name (such as "Hfo") that corresponds with the surface species name. The lists of surface species names and surface names are the same length. RM_FindComponents must be called before RM_GetSurfaceName. This method may be useful when generating selected output definitions related to surfaces. More... | |
| integer function | rm_getsurfacespeciescount (id) |
| Returns the number of surface species (such as "Hfo_wOH") in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSurfaceSpeciesCount. This method may be useful when generating selected output definitions related to surfaces. More... | |
| integer function | rm_getsurfacespeciesname (id, num, name) |
| Retrieves an item from the surface species list. The list of surface species (for example, "Hfo_wOH") is derived from the list of components (RM_FindComponents) and the list of all surface types (such as "Hfo_w") that are included in SURFACE definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSurfaceSpeciesName. This method may be useful when generating selected output definitions related to surfaces. More... | |
| integer function | rm_getsurfacetype (id, num, name) |
| Retrieves the surface site type (such as "Hfo_w") that corresponds with the surface species name. The lists of surface species names and surface species types are the same length. RM_FindComponents must be called before RM_GetSurfaceType. This method may be useful when generating selected output definitions related to surfaces. More... | |
| integer function | rm_getthreadcount (id) |
| Returns the number of threads, which is equal to the number of workers used to run in parallel with OPENMP. For the OPENMP version, the number of threads is set implicitly or explicitly with RM_Create. For the MPI version, the number of threads is always one for each process. More... | |
| double precision function | rm_gettime (id) |
| Returns the current simulation time in seconds. The reaction module does not change the time value, so the returned value is equal to the default (0.0) or the last time set by RM_SetTime. More... | |
| double precision function | rm_gettimeconversion (id) |
| Returns a multiplier to convert time from seconds to another unit, as specified by the user. The reaction module uses seconds as the time unit. The user can set a conversion factor (RM_SetTimeConversion) and retrieve it with RM_GetTimeConversion. The reaction module only uses the conversion factor when printing the long version of cell chemistry (RM_SetPrintChemistryOn), which is rare. Default conversion factor is 1.0. More... | |
| double precision function | rm_gettimestep (id) |
| Returns the current simulation time step in seconds. This is the time over which kinetic reactions are integrated in a call to RM_RunCells. The reaction module does not change the time step value, so the returned value is equal to the default (0.0) or the last time step set by RM_SetTimeStep. More... | |
| integer function | rm_initialphreeqc2concentrations (id, bc_conc, n_boundary, bc1, bc2, f1) |
| Fills an array (bc_conc) with concentrations from solutions in the InitialPhreeqc instance. The method is used to obtain concentrations for boundary conditions. If a negative value is used for a cell in bc1, then the highest numbered solution in the InitialPhreeqc instance will be used for that cell. Concentrations may be a mixture of two solutions, bc1 and bc2, with a mixing fraction for bc1 1 of f1 and mixing fraction for bc2 of (1 - f1). A negative value for bc2 implies no mixing, and the associated value for f1 is ignored. If bc2 and f1 are omitted, no mixing is used; concentrations are derived from bc1 only. More... | |
| integer function | rm_initialphreeqc2module (id, ic1, ic2, f1) |
| Transfer solutions and reactants from the InitialPhreeqc instance to the reaction-module workers, possibly with mixing. In its simplest form, ic1 is used to select initial conditions, including solutions and reactants, for each cell of the model, without mixing. ic1 is dimensioned (nxyz, 7), where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). The dimension of 7 refers to solutions and reactants in the following order: (1) SOLUTIONS, (2) EQUILIBRIUM_PHASES, (3) EXCHANGE, (4) SURFACE, (5) GAS_PHASE, (6) SOLID_SOLUTIONS, and (7) KINETICS. In Fortran, ic1(100, 4) = 2, indicates that cell 99 (0 based) contains the SURFACE definition with user number 2 that has been defined in the InitialPhreeqc instance (either by RM_RunFile or RM_RunString). It is also possible to mix solutions and reactants to obtain the initial conditions for cells. For mixing, ic2 contains numbers for a second entity that mixes with the entity defined in ic1. F1 contains the mixing fraction for ic1, whereas (1 - f1) is the mixing fraction for ic2. In Fortran, ic1(100, 4) = 2, initial_conditions2(100, 4) = 3, f1(100, 4) = 0.25 indicates that cell 99 (0 based) contains a mixture of 0.25 SURFACE 2 and 0.75 SURFACE 3, where the surface compositions have been defined in the InitialPhreeqc instance. If the user number in ic2 is negative, no mixing occurs. If ic2 and f1 are omitted, no mixing is used, and initial conditions are derived solely from ic1. More... | |
| integer function | rm_initialphreeqc2speciesconcentrations (id, bc_conc, n_boundary, bc1, bc2, f1) |
| Fills an array (bc_conc) with aqueous species concentrations from solutions in the InitialPhreeqc instance. This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The method is used to obtain aqueous species concentrations for boundary conditions. If a negative value is used for a cell in bc1, then the highest numbered solution in the InitialPhreeqc instance will be used for that cell. Concentrations may be a mixture of two solutions, bc1 and bc2, with a mixing fraction for bc1 of f1 and mixing fraction for bc2 of (1 - f1). A negative value for bc2 implies no mixing, and the associated value for f1 is ignored. If bc2 and f1 are omitted, no mixing is used; concentrations are derived from bc1 only. More... | |
| integer function | rm_initialphreeqccell2module (id, n_user, cell_numbers, n_cell) |
| A cell numbered n_user in the InitialPhreeqc instance is selected to populate a series of cells. All reactants with the number n_user are transferred along with the solution. If MIX n_user exists, it is used for the definition of the solution. If n_user is negative, n_user is redefined to be the largest solution or MIX number in the InitialPhreeqc instance. All reactants for each cell in the list cell_numbers are removed before the cell definition is copied from the InitialPhreeqc instance to the workers. More... | |
| integer function | rm_loaddatabase (id, db_name) |
| Load a database for all IPhreeqc instances–workers, InitialPhreeqc, and Utility. All definitions of the reaction module are cleared (SOLUTION_SPECIES, PHASES, SOLUTIONs, etc.), and the database is read. More... | |
| integer function | rm_logmessage (id, str) |
| Print a message to the log file. More... | |
| integer function | rm_mpiworker (id) |
| MPI only. Workers (processes with RM_GetMpiMyself > 0) must call RM_MpiWorker to be able to respond to messages from the root to accept data, perform calculations, and (or) return data. RM_MpiWorker contains a loop that reads a message from root, performs a task, and waits for another message from root. RM_SetConcentrations, RM_RunCells, and RM_GetConcentrations are examples of methods that send a message from root to get the workers to perform a task. The workers will respond to all methods that are designated "workers must be in the loop of RM_MpiWorker" in the MPI section of the method documentation. The workers will continue to respond to messages from root until root calls RM_MpiWorkerBreak. (Advanced) The list of tasks that the workers perform can be extended by using RM_SetMpiWorkerCallback. It is then possible to use the MPI processes to perform other developer-defined tasks, such as transport calculations, without exiting from the RM_MpiWorker loop. Alternatively, root calls RM_MpiWorkerBreak to allow the workers to continue past a call to RM_MpiWorker. The workers perform developer-defined calculations, and then RM_MpiWorker is called again to respond to requests from root to perform reaction-module tasks. More... | |
| integer function | rm_mpiworkerbreak (id) |
| MPI only. This method is called by root to force workers (processes with RM_GetMpiMyself > 0) to return from a call to RM_MpiWorker. RM_MpiWorker contains a loop that reads a message from root, performs a task, and waits for another message from root. The workers respond to all methods that are designated "workers must be in the loop of RM_MpiWorker" in the MPI section of the method documentation. The workers will continue to respond to messages from root until root calls RM_MpiWorkerBreak. More... | |
| integer function | rm_openfiles (id) |
| Opens the output and log files. Files are named prefix.chem.txt and prefix.log.txt based on the prefix defined by RM_SetFilePrefix. More... | |
| integer function | rm_outputmessage (id, str) |
| Print a message to the output file. More... | |
| integer function | rm_runcells (id) |
| Runs a reaction step for all of the cells in the reaction module. Normally, tranport concentrations are transferred to the reaction cells (RM_SetConcentrations) before reaction calculations are run. The length of time over which kinetic reactions are integrated is set by RM_SetTimeStep. Other properties that may need to be updated as a result of the transport calculations include porosity (RM_SetPorosity), saturation (RM_SetSaturation), temperature (RM_SetTemperature), and pressure (RM_SetPressure). More... | |
| integer function | rm_runfile (id, workers, initial_phreeqc, utility, chem_name) |
| Run a PHREEQC input file. The first three arguments determine which IPhreeqc instances will run the file–the workers, the InitialPhreeqc instance, and (or) the Utility instance. Input files that modify the thermodynamic database should be run by all three sets of instances. Files with SELECTED_OUTPUT definitions that will be used during the time-stepping loop need to be run by the workers. Files that contain initial conditions or boundary conditions should be run by the InitialPhreeqc instance. More... | |
| integer function | rm_runstring (id, workers, initial_phreeqc, utility, input_string) |
| Run a PHREEQC input string. The first three arguments determine which IPhreeqc instances will run the string–the workers, the InitialPhreeqc instance, and (or) the Utility instance. Input strings that modify the thermodynamic database should be run by all three sets of instances. Strings with SELECTED_OUTPUT definitions that will be used during the time-stepping loop need to be run by the workers. Strings that contain initial conditions or boundary conditions should be run by the InitialPhreeqc instance. More... | |
| integer function | rm_screenmessage (id, str) |
| Print message to the screen. More... | |
| integer function | rm_setcomponenth2o (id, tf) |
| Select whether to include H2O in the component list. The concentrations of H and O must be known accurately (8 to 10 significant digits) for the numerical method of PHREEQC to produce accurate pH and pe values. Because most of the H and O are in the water species, it may be more robust (require less accuracy in transport) to transport the excess H and O (the H and O not in water) and water. The default setting (true) is to include water, excess H, and excess O as components. A setting of false will include total H and total O as components. RM_SetComponentH2O must be called before RM_FindComponents. More... | |
| integer function | rm_setconcentrations (id, c) |
| Use the vector of concentrations (c) to set the moles of components in each reaction cell. The volume of water in a cell is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and reference volume (RM_SetRepresentativeVolume). The moles of each component are determined by the volume of water and per liter concentrations. If concentration units (RM_SetUnitsSolution) are mass fraction, the density (as specified by RM_SetDensity) is used to convert from mass fraction to per mass per liter. More... | |
| integer function | rm_setconcentrations1d (id, c) |
| integer function | rm_setcurrentselectedoutputusernumber (id, n_user) |
| Select the current selected output by user number. The user may define multiple SELECTED_OUTPUT data blocks for the workers. A user number is specified for each data block. The value of the argument n_user selects which of the SELECTED_OUTPUT definitions will be used for selected-output operations. More... | |
| integer function | rm_setdensity (id, density) |
| Set the density for each reaction cell. These density values are used when converting from transported mass fraction concentrations (RM_SetUnitsSolution) to produce per liter concentrations during a call to RM_SetConcentrations. They are also used when converting from module concentrations to transport concentrations of mass fraction (RM_GetConcentrations), if RM_UseSolutionDensityVolume is set to false. More... | |
| integer function | rm_setdumpfilename (id, dump_name) |
| Set the name of the dump file. It is the name used by RM_DumpModule. More... | |
| integer function | rm_seterrorhandlermode (id, mode) |
| Set the action to be taken when the reaction module encounters an error. Options are 0, return to calling program with an error return code (default); 1, throw an exception, in C++, the exception can be caught, for C and Fortran, the program will exit; or 2, attempt to exit gracefully. More... | |
| integer function | rm_seterroron (id, tf) |
| Set the property that controls whether error messages are generated and displayed. Messages include PHREEQC "ERROR" messages, and any messages written with RM_ErrorMessage. More... | |
| integer function | rm_setfileprefix (id, prefix) |
| Set the prefix for the output (prefix.chem.txt) and log (prefix.log.txt) files. These files are opened by RM_OpenFiles. More... | |
| integer function | rm_setgascompmoles (id, gas_moles) |
| Use the array of concentrations (gas_moles) to set the moles of gas components in each reaction cell. More... | |
| integer function | rm_setgasphasevolume (id, gas_volume) |
| Transfer volumes of gas phases from the array given in the argument list (gas_volume) to each reaction cell. The gas-phase volume affects the pressures calculated for fixed-volume gas phases. If a gas-phase volume is defined with this method for a GAS_PHASE in a cell, the gas phase is forced to be a fixed-volume gas phase. More... | |
| integer function | rm_setmpiworkercallback (id, fcn) |
| MPI only. Defines a callback function that allows additional tasks to be done by the workers. The method RM_MpiWorker contains a loop, where the workers receive a message (an integer), run a function corresponding to that integer, and then wait for another message. RM_SetMpiWorkerCallback allows the developer to add another function that responds to additional integer messages by calling developer-defined functions corresponding to those integers. RM_MpiWorker calls the callback function when the message number is not one of the PhreeqcRM message numbers. Messages are unique integer numbers. PhreeqcRM uses integers in a range beginning at 0. It is suggested that developers use message numbers starting at 1000 or higher for their tasks. The callback function calls a developer-defined function specified by the message number and then returns to RM_MpiWorker to wait for another message. For Fortran, the functions that are called from the callback function can use USE statements to find the data necessary to perform the tasks, and the only argument to the callback function is an integer message argument. RM_SetMpiWorkerCallback must be called by each worker before RM_MpiWorker is called. The motivation for this method is to allow the workers to perform other tasks, for instance, parallel transport calculations, within the structure of RM_MpiWorker. The callback function can be used to allow the workers to receive data, perform transport calculations, and (or) send results, without leaving the loop of RM_MpiWorker. Alternatively, it is possible for the workers to return from RM_MpiWorker by a call to RM_MpiWorkerBreak by root. The workers could then call subroutines to receive data, calculate transport, and send data, and then resume processing PhreeqcRM messages from root with another call to RM_MpiWorker. More... | |
| integer function | rm_setpartitionuzsolids (id, tf) |
| Sets the property for partitioning solids between the saturated and unsaturated parts of a partially saturated cell. More... | |
| integer function | rm_setporosity (id, por) |
| Set the porosity for each reaction cell. The volume of water in a reaction cell is the product of the porosity, the saturation (RM_SetSaturation), and the representative volume (RM_SetRepresentativeVolume). More... | |
| integer function | rm_setpressure (id, p) |
| Set the pressure for each reaction cell. Pressure effects are considered only in three of the databases distributed with PhreeqcRM: phreeqc.dat, Amm.dat, and pitzer.dat. More... | |
| integer function | rm_setprintchemistrymask (id, cell_mask) |
| Enable or disable detailed output for each reaction cell. Printing for a cell will occur only when the printing is enabled with RM_SetPrintChemistryOn and the cell_mask value is 1. More... | |
| integer function | rm_setprintchemistryon (id, workers, initial_phreeqc, utility) |
| Setting to enable or disable printing detailed output from reaction calculations to the output file for a set of cells defined by RM_SetPrintChemistryMask. The detailed output prints all of the output typical of a PHREEQC reaction calculation, which includes solution descriptions and the compositions of all other reactants. The output can be several hundred lines per cell, which can lead to a very large output file (prefix.chem.txt, RM_OpenFiles). For the worker instances, the output can be limited to a set of cells (RM_SetPrintChemistryMask) and, in general, the amount of information printed can be limited by use of options in the PRINT data block of PHREEQC (applied by using RM_RunFile or RM_RunString). Printing the detailed output for the workers is generally used only for debugging, and PhreeqcRM will run significantly faster when printing detailed output for the workers is disabled. More... | |
| integer function | rm_setrebalancebycell (id, method) |
| Set the load-balancing algorithm. PhreeqcRM attempts to rebalance the load of each thread or process such that each thread or process takes the same amount of time to run its part of a RM_RunCells calculation. Two algorithms are available; one uses individual times for each cell and accounts for cells that were not run because saturation was zero (default), and the other assigns an average time to all cells. The methods are similar, but limited testing indicates the default method performs better. More... | |
| integer function | rm_setrebalancefraction (id, f) |
| Sets the fraction of cells that are transferred among threads or processes when rebalancing. PhreeqcRM attempts to rebalance the load of each thread or process such that each thread or process takes the same amount of time to run its part of a RM_RunCells calculation. The rebalancing transfers cell calculations among threads or processes to try to achieve an optimum balance. RM_SetRebalanceFraction adjusts the calculated optimum number of cell transfers by a fraction from 0 to 1.0 to determine the actual number of cell transfers. A value of zero eliminates load rebalancing. A value less than 1.0 is suggested to slow the approach to the optimum cell distribution and avoid possible oscillations when too many cells are transferred at one iteration, requiring reverse transfers at the next iteration. Default is 0.5. More... | |
| integer function | rm_setrepresentativevolume (id, rv) |
| Set the representative volume of each reaction cell. By default the representative volume of each reaction cell is 1 liter. The volume of water in a reaction cell is determined by the procuct of the representative volume, the porosity (RM_SetPorosity), and the saturation (RM_SetSaturation). The numerical method of PHREEQC is more robust if the water volume for a reaction cell is within a couple orders of magnitude of 1.0. Small water volumes caused by small porosities and (or) small saturations (and (or) small representative volumes) may cause non-convergence of the numerical method. In these cases, a larger representative volume may help. Note that increasing the representative volume also increases the number of moles of the reactants in the reaction cell (minerals, surfaces, exchangers, and others), which are defined as moles per representative volume. More... | |
| integer function | rm_setsaturation (id, sat) |
| Set the saturation of each reaction cell. Saturation is a fraction ranging from 0 to 1. The volume of water in a cell is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and representative volume (RM_SetRepresentativeVolume). As a result of a reaction calculation, solution properties (density and volume) will change; the databases phreeqc.dat, Amm.dat, and pitzer.dat have the molar volume data to calculate these changes. The methods RM_GetDensity, RM_GetSolutionVolume, and RM_GetSaturation can be used to account for these changes in the succeeding transport calculation. RM_SetRepresentativeVolume should be called before initial conditions are defined for the reaction cells. More... | |
| integer function | rm_setscreenon (id, tf) |
| Set the property that controls whether messages are written to the screen. Messages include information about rebalancing during RM_RunCells, and any messages written with RM_ScreenMessage. More... | |
| integer function | rm_setselectedoutputon (id, tf) |
| Setting determines whether selected-output results are available to be retrieved with RM_GetSelectedOutput. 1 indicates that selected-output results will be accumulated during RM_RunCells and can be retrieved with RM_GetSelectedOutput; 0 indicates that selected-output results will not be accumulated during RM_RunCells. More... | |
| integer function | rm_setspeciessaveon (id, save_on) |
| Sets the value of the species-save property. This method enables use of PhreeqcRM with multicomponent-diffusion transport calculations. By default, concentrations of aqueous species are not saved. Setting the species-save property to 1 allows aqueous species concentrations to be retrieved with RM_GetSpeciesConcentrations, and solution compositions to be set with RM_SpeciesConcentrations2Module. RM_SetSpeciesSaveOn must be called before calls to RM_FindComponents. More... | |
| integer function | rm_settemperature (id, t) |
| Set the temperature for each reaction cell. If RM_SetTemperature is not called, worker solutions will have temperatures as defined by initial conditions (RM_InitialPhreeqc2Module and RM_InitialPhreeqcCell2Module). More... | |
| integer function | rm_settime (id, time) |
| Set current simulation time for the reaction module. More... | |
| integer function | rm_settimeconversion (id, conv_factor) |
| Set a factor to convert to user time units. Factor times seconds produces user time units. More... | |
| integer function | rm_settimestep (id, time_step) |
| Set current time step for the reaction module. This is the length of time over which kinetic reactions are integrated. More... | |
| integer function | rm_setunitsexchange (id, option) |
| Sets input units for exchangers. In PHREEQC input, exchangers are defined by moles of exchange sites (Mp). RM_SetUnitsExchange specifies how the number of moles of exchange sites in a reaction cell (Mc) is calculated from the input value (Mp). More... | |
| integer function | rm_setunitsgasphase (id, option) |
| Set input units for gas phases. In PHREEQC input, gas phases are defined by moles of component gases (Mp). RM_SetUnitsGasPhase specifies how the number of moles of component gases in a reaction cell (Mc) is calculated from the input value (Mp). More... | |
| integer function | rm_setunitskinetics (id, option) |
| Set input units for kinetic reactants. More... | |
| integer function | rm_setunitsppassemblage (id, option) |
| Set input units for pure phase assemblages (equilibrium phases). In PHREEQC input, equilibrium phases are defined by moles of each phase (Mp). RM_SetUnitsPPassemblage specifies how the number of moles of phases in a reaction cell (Mc) is calculated from the input value (Mp). More... | |
| integer function | rm_setunitssolution (id, option) |
| Solution concentration units used by the transport model. Options are 1, mg/L; 2 mol/L; or 3, mass fraction, kg/kgs. PHREEQC defines solutions by the number of moles of each element in the solution. To convert from mg/L to moles of element in the representative volume of a reaction cell, mg/L is converted to mol/L and multiplied by the solution volume, which is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and representative volume (RM_SetRepresentativeVolume). To convert from mol/L to moles of element in the representative volume of a reaction cell, mol/L is multiplied by the solution volume. To convert from mass fraction to moles of element in the representative volume of a reaction cell, kg/kgs is converted to mol/kgs, multiplied by density (RM_SetDensity) and multiplied by the solution volume. More... | |
| integer function | rm_setunitsssassemblage (id, option) |
| Set input units for solid-solution assemblages. In PHREEQC, solid solutions are defined by moles of each component (Mp). RM_SetUnitsSSassemblage specifies how the number of moles of solid-solution components in a reaction cell (Mc) is calculated from the input value (Mp). More... | |
| integer function | rm_setunitssurface (id, option) |
| Set input units for surfaces. In PHREEQC input, surfaces are defined by moles of surface sites (Mp). RM_SetUnitsSurface specifies how the number of moles of surface sites in a reaction cell (Mc) is calculated from the input value (Mp). More... | |
| integer function | rm_speciesconcentrations2module (id, species_conc) |
| Set solution concentrations in the reaction cells based on the vector of aqueous species concentrations (species_conc). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. The method determines the total concentration of a component by summing the molarities of the individual species times the stoichiometric coefficient of the element in each species. Solution compositions in the reaction cells are updated with these component concentrations. More... | |
| integer function | rm_usesolutiondensityvolume (id, tf) |
| Determines the volume and density to use when converting from the reaction-module concentrations to transport concentrations (RM_GetConcentrations). Two options are available to convert concentration units: (1) the density and solution volume calculated by PHREEQC are used, or (2) the specified density (RM_SetDensity) and solution volume are defined by the product of saturation (RM_SetSaturation), porosity (RM_SetPorosity), and representative volume (RM_SetRepresentativeVolume). Transport models that consider density-dependent flow will probably use the PHREEQC-calculated density and solution volume (default), whereas transport models that assume constant-density flow will probably use specified values of density and solution volume. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate density and solution volume: phreeqc.dat, Amm.dat, and pitzer.dat. Density is only used when converting to transport units of mass fraction. More... | |
| integer function | rm_warningmessage (id, warn_str) |
| Print a warning message to the screen and the log file. More... | |
Fortran Documentation for the geochemical reaction module PhreeqcRM.
| integer function phreeqcrm::rm_abort | ( | integer, intent(in) | id, |
| integer, intent(in) | irm_result, | ||
| character(len=*), intent(in) | err_str | ||
| ) |
Abort the program. irm_result will be interpreted as an IRM_RESULT value and decoded; err_str will be printed; and the reaction module will be destroyed. If using MPI, an MPI_Abort message will be sent before the reaction module is destroyed. If the id is an invalid instance, RM_Abort will return a value of IRM_BADINSTANCE, otherwise the program will exit with a return code of 4.
| id | The instance id returned from RM_Create. |
| irm_result | Integer treated as an IRM_RESULT return code. |
| err_str | String to be printed as an error message. |
| IRM_RESULT | Program will exit before returning unless id is an invalid reaction module id. |
iphreeqc_id = RM_Concentrations2Utility(id, c_well, 1, tc, p_atm)
string = "SELECTED_OUTPUT 5; -pH;RUN_CELLS; -cells 1"
status = RunString(iphreeqc_id, string)
if (status .ne. 0) status = RM_Abort(id, status, "IPhreeqc RunString failed")
| integer function phreeqcrm::rm_closefiles | ( | integer, intent(in) | id | ) |
Close the output and log files.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_CloseFiles(id)
| integer function phreeqcrm::rm_concentrations2utility | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(in) | c, | ||
| integer, intent(in) | n, | ||
| double precision, dimension(:), intent(in) | tc, | ||
| double precision, dimension(:), intent(in) | p_atm | ||
| ) |
N sets of component concentrations are converted to SOLUTIONs numbered 1-n in the Utility IPhreeqc. The solutions can be reacted and manipulated with the methods of IPhreeqc. If solution concentration units (RM_SetUnitsSolution) are per liter, one liter of solution is created in the Utility instance; if solution concentration units are mass fraction, one kilogram of solution is created in the Utility instance. The motivation for this method is the mixing of solutions in wells, where it may be necessary to calculate solution properties (pH for example) or react the mixture to form scale minerals. The code fragments below make a mixture of concentrations and then calculate the pH of the mixture.
| id | The instance id returned from RM_Create. |
| c | Array of concentrations to be made SOLUTIONs in Utility IPhreeqc, array size is (n, ncomps) where ncomps is the number of components (RM_GetComponentCount). |
| n | The number of sets of concentrations. |
| tc | Array of temperatures to apply to the SOLUTIONs, in degree C. Array of size n. |
| p_atm | Array of pressures to apply to the SOLUTIONs, in atm. Array of size n. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate (c_well(1,ncomps))
do i = 1, ncomps
c_well(1,i) = 0.5 * c(1,i) + 0.5 * c(10,i)
enddo
allocate(tc(1), p_atm(1))
tc(1) = 15.0
p_atm(1) = 3.0
iphreeqc_id = RM_Concentrations2Utility(id, c_well, 1, tc, p_atm)
string = "SELECTED_OUTPUT 5; -pH; RUN_CELLS; -cells 1"
status = RunString(iphreeqc_id, string)
status = SetCurrentSelectedOutputUserNumber(iphreeqc_id, 5)
status = GetSelectedOutputValue(iphreeqc_id, 1, 1, vtype, pH, svalue)

| integer function phreeqcrm::rm_create | ( | integer, intent(in) | nxyz, |
| integer, intent(in) | nthreads | ||
| ) |
Creates a reaction module. If the code is compiled with the preprocessor directive USE_OPENMP, the reaction module is multithreaded. If the code is compiled with the preprocessor directive USE_MPI, the reaction module will use MPI and multiple processes. If neither preprocessor directive is used, the reaction module will be serial (unparallelized).
| nxyz | The number of grid cells in the user's model. |
| nthreads | (or comm, MPI) When using OPENMP, the argument (nthreads) is the number of worker threads to be used. If nthreads <= 0, the number of threads is set equal to the number of processors of the computer. When using MPI, the argument (comm) is the MPI communicator to use within the reaction module. |
| Id | of the PhreeqcRM instance, negative is failure (See RM_DecodeError). |
nxyz = 40
#ifdef USE_MPI
id = RM_Create(nxyz, MPI_COMM_WORLD)
call MPI_Comm_rank(MPI_COMM_WORLD, mpi_myself, status)
if (status .ne. MPI_SUCCESS) then
stop "Failed to get mpi_myself"
endif
if (mpi_myself > 0) then
status = RM_MpiWorker(id)
status = RM_Destroy(id)
return
endif
#else
nthreads = 3
id = RM_Create(nxyz, nthreads)
#endif

| integer function phreeqcrm::rm_createmapping | ( | integer, intent(in) | id, |
| integer, dimension(:), intent(in) | grid2chem | ||
| ) |
Provides a mapping from grid cells in the user's model to reaction cells in PhreeqcRM. The mapping is used to eliminate inactive cells and to use symmetry to decrease the number of cells for which chemistry must be run. The array grid2chem of size nxyz (the number of grid cells, RM_GetGridCellCount) must contain the set of all integers 0 <= i < count_chemistry, where count_chemistry is a number less than or equal to nxyz. Inactive cells are assigned a negative integer. The mapping may be many-to-one to account for symmetry. Default is a one-to-one mapping–all user grid cells are reaction cells (equivalent to grid2chem values of 0,1,2,3,...,nxyz-1).
| id | The instance id returned from RM_Create. |
| grid2chem | An array of integers: Nonnegative is a reaction cell number (0 based), negative is an inactive cell. Array of size nxyz (number of grid cells). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
! For demonstation, two equivalent rows by symmetry
allocate(grid2chem(nxyz))
do i = 1, nxyz/2
grid2chem(i) = i - 1
grid2chem(i+nxyz/2) = i - 1
enddo
status = RM_CreateMapping(id, grid2chem)

| integer function phreeqcrm::rm_decodeerror | ( | integer, intent(in) | id, |
| integer, intent(in) | e | ||
| ) |
If e is negative, this method prints an error message corresponding to IRM_RESULT e. If e is non-negative, no action is taken.
| id | The instance id returned from RM_Create. |
| e | An IRM_RESULT value returned by one of the reaction-module methods. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
typedef enum {
IRM_OK = 0, //Success
IRM_OUTOFMEMORY = -1, //Failure, Out of memory
IRM_BADVARTYPE = -2, //Failure, Invalid VAR type
IRM_INVALIDARG = -3, //Failure, Invalid argument
IRM_INVALIDROW = -4, //Failure, Invalid row
IRM_INVALIDCOL = -5, //Failure, Invalid column
IRM_BADINSTANCE = -6, //Failure, Invalid rm instance id
IRM_FAIL = -7, //Failure, Unspecified
} IRM_RESULT;
status = RM_CreateMapping(id, grid2chem)
if (status < 0) status = RM_DecodeError(id, status)
| integer function phreeqcrm::rm_destroy | ( | integer, intent(in) | id | ) |
Destroys a reaction module.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_Destroy(id)
| integer function phreeqcrm::rm_dumpmodule | ( | integer, intent(in) | id, |
| integer, intent(in) | dump_on, | ||
| integer, intent(in) | append | ||
| ) |
Writes the contents of all workers to file in _RAW formats, including SOLUTIONs and all reactants.
| id | The instance id returned from RM_Create. |
| dump_on | Signal for writing the dump file: 1 true, 0 false. |
| append | Signal to append to the contents of the dump file: 1 true, 0 false. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
dump_on = 1
append = 0
status = RM_SetDumpFileName(id, "advection_f90.dmp")
status = RM_DumpModule(id, dump_on, append)
| integer function phreeqcrm::rm_errormessage | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | errstr | ||
| ) |
Send an error message to the screen, the output file, and the log file.
| id | The instance id returned from RM_Create. |
| errstr | String to be printed. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_ErrorMessage(id, "Goodbye world")

| integer function phreeqcrm::rm_findcomponents | ( | integer, intent(in) | id | ) |
Returns the number of items in the list of all elements in the InitialPhreeqc instance. Elements are those that have been defined in a solution or any other reactant (EQUILIBRIUM_PHASE, KINETICS, and others). The method can be called multiple times and the list that is created is cummulative. The list is the set of components that needs to be transported. By default the list includes water, excess H and excess O (the H and O not contained in water); alternatively, the list may be set to contain total H and total O (RM_SetComponentH2O), which requires transport results to be accurate to eight or nine significant digits. If multicomponent diffusion (MCD) is to be modeled, there is a capability to retrieve aqueous species concentrations (RM_GetSpeciesConcentrations) and to set new solution concentrations after MCD by using individual species concentrations (RM_SpeciesConcentrations2Module). To use these methods the save-species property needs to be turned on (RM_SetSpeciesSaveOn). If the save-species property is on, RM_FindComponents will generate a list of aqueous species (RM_GetSpeciesCount, RM_GetSpeciesName), their diffusion coefficients at 25 C (RM_GetSpeciesD25), their charge (RM_GetSpeciesZ).
| id | The instance id returned from RM_Create. |
| Number | of components currently in the list, or IRM_RESULT error code (see RM_DecodeError). |
! Get list of components
ncomps = RM_FindComponents(id)
allocate(components(ncomps))
do i = 1, ncomps
status = RM_GetComponent(id, i, components(i))
enddo
| integer function phreeqcrm::rm_getbackwardmapping | ( | integer, intent(in) | id, |
| integer, intent(in) | n, | ||
| integer, dimension(*), intent(in) | list, | ||
| integer, intent(inout) | size | ||
| ) |
Fills an array with the cell numbers in the user's numbering sytstem that map to a cell in the PhreeqcRM numbering system. The mapping is defined by RM_CreateMapping.
| id | The instance id returned from RM_Create. |
| n | A cell number in the PhreeqcRM numbering system (0 <= n < RM_GetChemistryCellCount). |
| list | Array to store the user cell numbers mapped to PhreeqcRM cell n. |
| size | Input, the allocated size of list; it is an error if the array is too small. Output, the number of cells mapped to cell n. |
| IRM_RESULT | error code (see RM_DecodeError). |
if (RM_GetBackwardMapping(rm_id, rm_cell_number, list, size) .eq. 0) then
if (fstr(1:l) .eq. "HYDRAULIC_K") then
my_basic_fortran_callback = K_ptr(list(1)+1)
endif
endif
| integer function phreeqcrm::rm_getchemistrycellcount | ( | integer, intent(in) | id | ) |
Returns the number of chemistry cells in the reaction module. The number of chemistry cells is defined by the set of non-negative integers in the mapping from user grid cells (RM_CreateMapping). The number of chemistry cells is less than or equal to the number of cells in the user's model.
| id | The instance id returned from RM_Create. |
| Number | of chemistry cells, or IRM_RESULT error code (see RM_DecodeError). |
status = RM_CreateMapping(id, grid2chem)
nchem = RM_GetChemistryCellCount(id)
| integer function phreeqcrm::rm_getcomponent | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | comp_name | ||
| ) |
Retrieves an item from the reaction-module component list that was generated by calls to RM_FindComponents.
| id | The instance id returned from RM_Create. |
| num | The number of the component to be retrieved. Fortran, 1 based. |
| comp_name | The string value associated with component num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
! Get list of components
ncomps = RM_FindComponents(id)
allocate(components(ncomps))
do i = 1, ncomps
status = RM_GetComponent(id, i, components(i))
enddo
| integer function phreeqcrm::rm_getcomponentcount | ( | integer, intent(in) | id | ) |
Returns the number of components in the reaction-module component list. The component list is generated by calls to RM_FindComponents. The return value from the last call to RM_FindComponents is equal to the return value from RM_GetComponentCount.
| id | The instance id returned from RM_Create. |
| The | number of components in the reaction-module component list, negative is failure (See RM_DecodeError). |
ncomps1 = RM_GetComponentCount(id)
| integer function phreeqcrm::rm_getconcentrations | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | c | ||
| ) |
Transfer solution concentrations from each reaction cell to the concentration array given in the argument list (c). Units of concentration for c are defined by RM_SetUnitsSolution. For concentration units of per liter, the solution volume is used to calculate the concentrations for c. For mass fraction concentration units, the solution mass is used to calculate concentrations for c. Two options are available for the volume and mass of solution that are used in converting to transport concentrations: (1) the volume and mass of solution are calculated by PHREEQC, or (2) the volume of solution is the product of saturation (RM_SetSaturation), porosity (RM_SetPorosity), and representative volume (RM_SetRepresentativeVolume), and the mass of solution is volume times density as defined by RM_SetDensity. RM_UseSolutionDensityVolume determines which option is used. For option 1, the databases that have partial molar volume definitions needed to accurately calculate solution volume are phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| c | Array to receive the concentrations. Dimension of the array is (nxyz, ncomps), where nxyz is the number of user grid cells and ncomps is the result of RM_FindComponents or RM_GetComponentCount. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(c(nxyz, ncomps))
status = RM_RunCells(id)
status = RM_GetConcentrations(id, c)

| integer function phreeqcrm::rm_getdensity | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out) | density | ||
| ) |
Transfer solution densities from the reaction cells to the array given in the argument list (density). Densities are those calculated by the reaction module. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate density: phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| density | Array to receive the densities. Dimension of the array is nxyz, where nxyz is the number of user grid cells (RM_GetGridCellCount). Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(density(nxyz))
status = RM_RunCells(id)
status = RM_GetDensity(id, density)

| integer function phreeqcrm::rm_getendcell | ( | integer, intent(in) | id, |
| integer, dimension(:), intent(out) | ec | ||
| ) |
Returns an array with the ending cell numbers from the range of cell numbers assigned to each worker.
| id | The instance id returned from RM_Create. |
| ec | Array to receive the ending cell numbers. Dimension of the array is the number of threads (OpenMP) or the number of processes (MPI). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
n = RM_GetThreadCount(id) * RM_GetMpiTasks(id)
allocate(ec(n))
status = RM_GetEndCell(id, ec)
| integer function phreeqcrm::rm_getequilibriumphasescount | ( | integer, intent(in) | id | ) |
Returns the number of equilibrium phases in the initial-phreeqc module. RM_FindComponents must be called before RM_GetEquilibriumPhasesCount. This method may be useful when generating selected output definitions related to equilibrium phases.
| id | The instance id returned from RM_Create. |
| The | number of equilibrium phases in the initial-phreeqc module. |
input = trim(input) // " -equilibrium_phases " // new_line(c)
do i = 1, RM_GetEquilibriumPhasesCount(id)
status = RM_GetEquilibriumPhasesName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getequilibriumphasesname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the equilibrium phase list. The list includes all phases included in any EQUILIBRIUM_PHASES definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetEquilibriumPhasesName. This method may be useful when generating selected output definitions related to equilibrium phases.
| id | The instance id returned from RM_Create. |
| num | The number of the equilibrium phase name to be retrieved. Fortran, 1 based. |
| name | The equilibrium phase name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
input = trim(input) // " -equilibrium_phases " // new_line(c)
do i = 1, RM_GetEquilibriumPhasesCount(id)
status = RM_GetEquilibriumPhasesName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_geterrorstring | ( | integer, intent(in) | id, |
| character(len=*), intent(out) | errstr | ||
| ) |
Returns a string containing error messages related to the last call to a PhreeqcRM method to the character argument (errstr).
| id | The instance id returned from RM_Create. |
| errstr | The error string related to the last call to a PhreeqcRM method. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
character(len=:), allocatable :: errstr
integer l
if (status .ne. 0) then
l = RM_GetErrorStringLength(id)
allocate (character(len=l) :: errstr)
status = RM_GetErrorString(id, errstr)
write(*,"(A)") errstr
deallocate(errstr)
status = RM_Destroy(id)
stop
endif
| integer function phreeqcrm::rm_geterrorstringlength | ( | integer, intent(in) | id | ) |
Returns the length of the string that contains error messages related to the last call to a PhreeqcRM method.
| id | The instance id returned from RM_Create. |
| int | Length of the error message string. |
character(len=:), allocatable :: errstr
integer l
if (status .ne. 0) then
l = RM_GetErrorStringLength(id)
allocate (character(len=l) :: errstr)
status = RM_GetErrorString(id, errstr)
write(*,"(A)") errstr
deallocate(errstr)
status = RM_Destroy(id)
stop
endif
| integer function phreeqcrm::rm_getexchangename | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the exchange name list. RM_FindComponents must be called before RM_GetExchangeName. The exchange names vector is the same length as the exchange species names vector and provides the corresponding exchange site (for example, X corresponing to NaX). This method may be useful when generating selected output definitions related to exchangers.
| id | The instance id returned from RM_Create. |
| num | The number of the exchange name to be retrieved. Fortran, 1 based. |
| name | The exchange name associated with exchange species num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do i = 1, RM_GetExchangeSpeciesCount(id)
status = RM_GetExchangeSpeciesName(id, i, line)
status = RM_GetExchangeName(id, i, line1)
input = trim(input) // " " // line // " # " // line1 // new_line(c)
enddo
| integer function phreeqcrm::rm_getexchangespeciescount | ( | integer, intent(in) | id | ) |
Returns the number of exchange species in the initial-phreeqc module. RM_FindComponents must be called before RM_GetExchangeSpeciesCount. This method may be useful when generating selected output definitions related to exchangers.
| id | The instance id returned from RM_Create. |
| The | number of exchange species in the initial-phreeqc module. |
do i = 1, RM_GetExchangeSpeciesCount(id)
status = RM_GetExchangeSpeciesName(id, i, line)
status = RM_GetExchangeName(id, i, line1)
input = trim(input) // " " // line // " # " // line1 // new_line(c)
enddo
| integer function phreeqcrm::rm_getexchangespeciesname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the exchange species list. The list of exchange species (such as "NaX") is derived from the list of components (RM_FindComponents) and the list of all exchange names (such as "X") that are included in EXCHANGE definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetExchangeSpeciesName. This method may be useful when generating selected output definitions related to exchangers.
| id | The instance id returned from RM_Create. |
| num | The number of the exchange species to be retrieved. Fortran, 1 based. |
| name | The exchange species name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do i = 1, RM_GetExchangeSpeciesCount(id)
status = RM_GetExchangeSpeciesName(id, i, line)
status = RM_GetExchangeName(id, i, line1)
input = trim(input) // " " // line // " # " // line1 // new_line(c)
enddo
| integer function phreeqcrm::rm_getfileprefix | ( | integer, intent(in) | id, |
| character(len=*), intent(inout) | prefix | ||
| ) |
Returns the reaction-module file prefix to the character argument (prefix).
| id | The instance id returned from RM_Create. |
| prefix | Character string where the prefix is written. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
character(100) :: string
character(200) :: string1
status = RM_GetFilePrefix(id, string)
string1 = "File prefix: "//string
status = RM_OutputMessage(id, string1)
| integer function phreeqcrm::rm_getgascompmoles | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out), target | gas_moles | ||
| ) |
Transfer moles of gas components from each reaction cell to the array given in the argument list (gas_moles).
| id | The instance id returned from RM_Create. |
| gas_moles | Array to receive the moles of gas components for each cell. Dimension of the array is (nxyz, ngas_comps), where nxyz is the number of user grid cells and ngas_comps is the result of RM_GetGasComponentsCount. If a gas component is not defined for a cell, the number of moles is set to -1. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
ngas_comps = RM_GetGasComponentsCount(id)
allocate(gas_moles(nxyz, ngas_comps))
status = RM_RunCells(id)
status = RM_GetGasCompMoles(id, gas_moles)
| integer function phreeqcrm::rm_getgascomponentscount | ( | integer, intent(in) | id | ) |
Returns the number of gas phase components in the initial-phreeqc module. RM_FindComponents must be called before RM_GetGasComponentsCount. This method may be useful when generating selected output definitions related to gas phases.
| id | The instance id returned from RM_Create. |
| The | number of gas phase components in the initial-phreeqc module. |
input = trim(input) // " -gas " // new_line(c)
do i = 1, RM_GetGasComponentsCount(id)
status = RM_GetGasComponentsName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getgascomponentsname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the gas components list. The list includes all gas components included in any GAS_PHASE definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetGasComponentsName. This method may be useful when generating selected output definitions related to gas phases.
| id | The instance id returned from RM_Create. |
| num | The number of the gas component name to be retrieved. Fortran, 1 based. |
| name | The gas component name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
input = trim(input) // " -gas " // new_line(c)
do i = 1, RM_GetGasComponentsCount(id)
status = RM_GetGasComponentsName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getgascompphi | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out), target | gas_phi | ||
| ) |
Transfer fugacity coefficients (phi) of gas components from each reaction cell to the array given in the argument list (gas_phi). Fugacity of a gas component is equal to the pressure of the component times the fugacity coefficient.
| id | The instance id returned from RM_Create. |
| gas_phi | Array to receive the fugacity coefficients of gas components for each cell. Dimension of the array is (nxyz, ngas_comps), where nxyz is the number of user grid cells and ngas_comps is the result of RM_GetGasComponentsCount. If a gas component is not defined for a cell, the fugacity coefficient is set to -1. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
ngas_comps = RM_GetGasComponentsCount(id)
allocate(gas_phi(nxyz, ngas_comps))
status = RM_RunCells(id)
status = RM_GetGasCompPhi(id, gas_phi)
| integer function phreeqcrm::rm_getgascomppressures | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out), target | gas_p | ||
| ) |
Transfer pressures of gas components from each reaction cell to the array given in the argument list (gas_p).
| id | The instance id returned from RM_Create. |
| gas_p | Array to receive the moles of gas components for each cell. Dimension of the array is (nxyz, ngas_comps), where nxyz is the number of user grid cells and ngas_comps is the result of RM_GetGasComponentsCount. If a gas component is not defined for a cell, the pressure is set to -1. Values for inactive cells are set to 1e30. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
ngas_comps = RM_GetGasComponentsCount(id)
allocate(gas_p(nxyz, ngas_comps))
status = RM_RunCells(id)
status = RM_GetGasCompPressures(id, gas_p)
| integer function phreeqcrm::rm_getgasphasevolume | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out), target | gas_volume | ||
| ) |
Transfer volume of gas from each reaction cell to the vector given in the argument list (gas_volume).
| id | The instance id returned from RM_Create. |
| gas_volume | Array to receive the gas phase volumes. Dimension of the array must be nxyz, where nxyz is the number of user grid cells (RM_GetGridCellCount). If a gas phase is not defined for a cell, the volume is set to -1. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
ngas_comps = RM_GetGasComponentsCount(id)
allocate(gas_volume(nxyz, ngas_comps))
status = RM_RunCells(id)
status = RM_GetGasPhaseVolume(id, gas_volume)
| integer function phreeqcrm::rm_getgfw | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out) | gfw | ||
| ) |
Returns the gram formula weights (g/mol) for the components in the reaction-module component list.
| id | The instance id returned from RM_Create. |
| gfw | Array to receive the gram formula weights. Dimension of the array is ncomps, where ncomps is the number of components in the component list. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
character(100), dimension(:), allocatable :: components
double precision, dimension(:), allocatable :: gfw
ncomps = RM_FindComponents(id)
allocate(components(ncomps))
allocate(gfw(ncomps))
status = RM_GetGfw(id, gfw)
do i = 1, ncomps
status = RM_GetComponent(id, i, components(i))
write(string,"(A10, F15.4)") components(i), gfw(i)
status = RM_OutputMessage(id, string)
enddo

| integer function phreeqcrm::rm_getgridcellcount | ( | integer, intent(in) | id | ) |
Returns the number of grid cells in the user's model, which is defined in the call to RM_Create. The mapping from grid cells to reaction cells is defined by RM_CreateMapping. The number of reaction cells may be less than the number of grid cells if there are inactive regions or symmetry in the model definition.
| id | The instance id returned from RM_Create. |
| Number | of grid cells in the user's model, negative is failure (See RM_DecodeError). |
nxyz = RM_GetGridCellCount(id)
write(string1, "(A,I)") "Number of grid cells in the user's model: ", nxyz
status = RM_OutputMessage(id, trim(string1))
| integer function phreeqcrm::rm_getiphreeqcid | ( | integer, intent(in) | id, |
| integer, intent(in) | i | ||
| ) |
Returns an IPhreeqc id for the ith IPhreeqc instance in the reaction module. For the threaded version, there are nthreads + 2 IPhreeqc instances, where nthreads is defined in the constructor (RM_Create). The number of threads can be determined by RM_GetThreadCount. The first nthreads (0 based) instances will be the workers, the next (nthreads) is the InitialPhreeqc instance, and the next (nthreads + 1) is the Utility instance. Getting the IPhreeqc pointer for one of these instances allows the user to use any of the IPhreeqc methods on that instance. For MPI, each process has exactly three IPhreeqc instances, one worker (number 0), one InitialPhreeqc instance (number 1), and one Utility instance (number 2).
| id | The instance id returned from RM_Create. |
| i | The number of the IPhreeqc instance to be retrieved (0 based). |
| IPhreeqc | id for the ith IPhreeqc instance, negative is failure (See RM_DecodeError). |
! Utility pointer is worker number nthreads + 1
iphreeqc_id1 = RM_GetIPhreeqcId(id, RM_GetThreadCount(id) + 1)
| integer function phreeqcrm::rm_getkineticreactionscount | ( | integer, intent(in) | id | ) |
Returns the number of kinetic reactions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetKineticReactionsCount. This method may be useful when generating selected output definitions related to kinetic reactions.
| id | The instance id returned from RM_Create. |
| The | number of kinetic reactions in the initial-phreeqc module. |
input = trim(input) // " -kinetics " // new_line(c)
do i = 1, RM_GetKineticReactionsCount(id)
status = RM_GetKineticReactionsName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getkineticreactionsname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the kinetic reactions list. The list includes all kinetic reactions included in any KINETICS definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetKineticReactionsName. This method may be useful when generating selected output definitions related to kinetic reactions.
| id | The instance id returned from RM_Create. |
| num | The number of the kinetic reaction name to be retrieved. Fortran, 1 based. |
| name | The kinetic reaction name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
input = trim(input) // " -kinetics " // new_line(c)
do i = 1, RM_GetKineticReactionsCount(id)
status = RM_GetKineticReactionsName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getmpimyself | ( | integer, intent(in) | id | ) |
Returns the MPI task number. For the OPENMP version, the task number is always zero and the result of RM_GetMpiTasks is one. For the MPI version, the root task number is zero, and all workers have a task number greater than zero. The number of tasks can be obtained with RM_GetMpiTasks. The number of tasks and computer hosts are determined at run time by the mpiexec command, and the number of reaction-module processes is defined by the communicator used in constructing the reaction modules (RM_Create).
| id | The instance id returned from RM_Create. |
| The | MPI task number for a process, negative is failure (See RM_DecodeError). |
write(string1, "(A,I)") "MPI task number: ", RM_GetMpiMyself(id)
status = RM_OutputMessage(id, string1)
| integer function phreeqcrm::rm_getmpitasks | ( | integer, intent(in) | id | ) |
Returns the number of MPI processes (tasks) assigned to the reaction module. For the OPENMP version, the number of tasks is always one (although there may be multiple threads, RM_GetThreadCount), and the task number returned by RM_GetMpiMyself is zero. For the MPI version, the number of tasks and computer hosts are determined at run time by the mpiexec command. An MPI communicator is used in constructing reaction modules for MPI. The communicator may define a subset of the total number of MPI processes. The root task number is zero, and all workers have a task number greater than zero.
| id | The instance id returned from RM_Create. |
| The | number of MPI processes assigned to the reaction module, negative is failure (See RM_DecodeError). |
mpi_tasks = RM_GetMpiTasks(id)
write(string1, "(A,I)") "Number of MPI processes: ", mpi_tasks
status = RM_OutputMessage(id, string1)
| integer function phreeqcrm::rm_getnthselectedoutputusernumber | ( | integer, intent(in) | id, |
| integer, intent(in) | n | ||
| ) |
Returns the user number for the nth selected-output definition. Definitions are sorted by user number. Phreeqc allows multiple selected-output definitions, each of which is assigned a nonnegative integer identifier by the user. The number of definitions can be obtained by RM_GetSelectedOutputCount. To cycle through all of the definitions, RM_GetNthSelectedOutputUserNumber can be used to identify the user number for each selected-output definition in sequence. RM_SetCurrentSelectedOutputUserNumber is then used to select that user number for selected-output processing.
| id | The instance id returned from RM_Create. |
| n | The sequence number of the selected-output definition for which the user number will be returned. Fortran, 1 based. |
| The | user number of the nth selected-output definition, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
write(*,*) "Selected output sequence number: ", isel)
write(*,*) "Selected output user number: ", n_user)
col = RM_GetSelectedOutputColumnCount(id)
allocate(selected_out(nxyz,col))
status = RM_GetSelectedOutput(id, selected_out)
! Process results here
deallocate(selected_out)
enddo
| integer function phreeqcrm::rm_getsaturation | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out) | sat_calc | ||
| ) |
Returns a vector of saturations (sat_calc) as calculated by the reaction module. Reactions will change the volume of solution in a cell. The transport code must decide whether to ignore or account for this change in solution volume due to reactions. Following reactions, the cell saturation is calculated as solution volume (RM_GetSolutionVolume) divided by the product of representative volume (RM_SetRepresentativeVolume) and the porosity (RM_SetPorosity). The cell saturation returned by RM_GetSaturation may be less than or greater than the saturation set by the transport code (RM_SetSaturation), and may be greater than or less than 1.0, even in fully saturated simulations. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate solution volume and saturation: phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| sat_calc | Vector to receive the saturations. Dimension of the array is set to nxyz, where nxyz is the number of user grid cells (RM_GetGridCellCount). Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(sat_calc(nxyz))
status = RM_RunCells(id)
status = RM_GetSaturation(id, sat_calc)

| integer function phreeqcrm::rm_getselectedoutput | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | so | ||
| ) |
Populates an array with values from the current selected-output definition. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used to populate the array.
| id | The instance id returned from RM_Create. |
| so | An array to contain the selected-output values. Size of the array is (nxyz, col), where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount), and col is the number of columns in the selected-output definition (RM_GetSelectedOutputColumnCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
col = RM_GetSelectedOutputColumnCount(id)
allocate(selected_out(nxyz,col))
status = RM_GetSelectedOutput(id, selected_out)
! Process results here
deallocate(selected_out)
enddo
| integer function phreeqcrm::rm_getselectedoutputcolumncount | ( | integer, intent(in) | id | ) |
Returns the number of columns in the current selected-output definition. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used.
| id | The instance id returned from RM_Create. |
| Number | of columns in the current selected-output definition, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
col = RM_GetSelectedOutputColumnCount(id)
allocate(selected_out(nxyz,col))
status = RM_GetSelectedOutput(id, selected_out)
! Process results here
deallocate(selected_out)
enddo

| integer function phreeqcrm::rm_getselectedoutputcount | ( | integer, intent(in) | id | ) |
Returns the number of selected-output definitions. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used.
| id | The instance id returned from RM_Create. |
| Number | of selected-output definitions, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
col = RM_GetSelectedOutputColumnCount(id)
allocate(selected_out(nxyz,col))
status = RM_GetSelectedOutput(id, selected_out)
! Process results here
deallocate(selected_out)
enddo
| integer function phreeqcrm::rm_getselectedoutputheading | ( | integer, intent(in) | id, |
| integer, intent(in) | icol, | ||
| character(len=*), intent(out) | heading | ||
| ) |
Returns a selected output heading. The number of headings is determined by RM_GetSelectedOutputColumnCount. RM_SetCurrentSelectedOutputUserNumber determines which of the selected-output definitions is used.
| id | The instance id returned from RM_Create. |
| icol | The sequence number of the heading to be retrieved. Fortran, 1 based. |
| heading | A string buffer to receive the heading. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
col = RM_GetSelectedOutputColumnCount(id)
do j = 1, col
status = RM_GetSelectedOutputHeading(id, j, heading)
write(*,'(10x,i2,A2,A10,A2,f10.4)') j, " ", trim(heading)
enddo
enddo
| integer function phreeqcrm::rm_getselectedoutputrowcount | ( | integer, intent(in) | id | ) |
Returns the number of rows in the current selected-output definition. However, the method is included only for convenience; the number of rows is always equal to the number of grid cells in the user's model, and is equal to RM_GetGridCellCount.
| id | The instance id returned from RM_Create. |
| Number | of rows in the current selected-output definition, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
col = RM_GetSelectedOutputColumnCount(id)
allocate(selected_out(nxyz,col))
status = RM_GetSelectedOutput(id, selected_out)
! Print results
do i = 1, RM_GetSelectedOutputRowCount(id)
write(*,*) "Cell number ", i
write(*,*) " Selected output: "
do j = 1, col
status = RM_GetSelectedOutputHeading(id, j, heading)
write(*,'(10x,i2,A2,A10,A2,f10.4)') j, " ", trim(heading),": ", selected_out(i,j)
enddo
enddo
deallocate(selected_out)
enddo
| integer function phreeqcrm::rm_getsicount | ( | integer, intent(in) | id | ) |
Returns the number of phases in the initial-phreeqc module for which saturation indices can be calculated. RM_FindComponents must be called before RM_GetSICount. This method may be useful when generating selected output definitions related to saturation indices.
| id | The instance id returned from RM_Create. |
| The | number of phases in the initial-phreeqc module for which saturation indices could be calculated. |
input = trim(input) // " -si "// new_line(c)
do i = 1, RM_GetSICount(id)
status = RM_GetSIName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getsiname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the list of all phases for which saturation indices can be calculated. The list includes all phases that contain only elements included in the components in the initial-phreeqc module. The list assumes that all components are present to be able to calculate the entire list of SIs; it may be that one or more components are missing in any specific cell. RM_FindComponents must be called before RM_GetSIName. This method may be useful when generating selected output definitions related to saturation indices.
| id | The instance id returned from RM_Create. |
| num | The number of the saturation-index-phase name to be retrieved. Fortran, 1 based. |
| name | The saturation-index-phase name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
input = trim(input) // " -si "// new_line(c)
do i = 1, RM_GetSICount(id)
status = RM_GetSIName(id, i, line)
input = trim(input) // " " // line // new_line(c)
enddo
| integer function phreeqcrm::rm_getsolidsolutioncomponentscount | ( | integer, intent(in) | id | ) |
Returns the number of solid solution components in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSolidSolutionComponentsCount. This method may be useful when generating selected output definitions related to solid solutions.
| id | The instance id returned from RM_Create. |
| The | number of solid solution components in the initial-phreeqc module. |
input = trim(input) // " -solid_solutions " // new_line(c)
do i = 1, RM_GetSolidSolutionComponentsCount(id)
status = RM_GetSolidSolutionComponentsName(id, i, line)
status = RM_GetSolidSolutionName(id, i, line1)
input = trim(input) // " " // line // " # " // line1 // new_line(c)
enddo
| integer function phreeqcrm::rm_getsolidsolutioncomponentsname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the solid solution components list. The list includes all solid solution components included in any SOLID_SOLUTIONS definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSolidSolutionComponentsName. This method may be useful when generating selected output definitions related to solid solutions.
| id | The instance id returned from RM_Create. |
| num | The number of the solid solution components name to be retrieved. Fortran, 1 based. |
| name | The solid solution compnent name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
input = trim(input) // " -solid_solutions " // new_line(c)
do i = 1, RM_GetSolidSolutionComponentsCount(id)
status = RM_GetSolidSolutionComponentsName(id, i, line)
status = RM_GetSolidSolutionName(id, i, line1)
input = trim(input) // " " // line // " # " // line1 // new_line(c)
enddo
| integer function phreeqcrm::rm_getsolidsolutionname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the solid solution names list. The list includes solid solution names included in SOLID_SOLUTIONS definitions in the initial-phreeqc module. The solid solution names vector is the same length as the solid solution components vector and provides the corresponding name of solid solution containing the component. RM_FindComponents must be called before RM_GetSolidSolutionName. This method may be useful when generating selected output definitions related to solid solutions.
| id | The instance id returned from RM_Create. |
| num | The number of the solid solution name to be retrieved. Fortran, 1 based. |
| name | The solid solution name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
input = trim(input) // " -solid_solutions " // new_line(c)
do i = 1, RM_GetSolidSolutionComponentsCount(id)
status = RM_GetSolidSolutionComponentsName(id, i, line)
status = RM_GetSolidSolutionName(id, i, line1)
input = trim(input) // " " // line // " # " // line1 // new_line(c)
enddo
| integer function phreeqcrm::rm_getsolutionvolume | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out) | vol | ||
| ) |
Transfer solution volumes from the reaction cells to the array given in the argument list (vol). Solution volumes are those calculated by the reaction module. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate solution volume: phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| vol | Array to receive the solution volumes. Dimension of the array is (nxyz), where nxyz is the number of user grid cells. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(volume(nxyz))
status = RM_RunCells(id)
status = RM_GetSolutionVolume(id, volume)

| integer function phreeqcrm::rm_getspeciesconcentrations | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | species_conc | ||
| ) |
Transfer concentrations of aqueous species to the array argument (species_conc) This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. Solution volumes used to calculate mol/L are calculated by the reaction module. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate solution volume: phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| species_conc | Array to receive the aqueous species concentrations. Dimension of the array is (nxyz, nspecies), where nxyz is the number of user grid cells (RM_GetGridCellCount), and nspecies is the number of aqueous species (RM_GetSpeciesCount). Concentrations are moles per liter. Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
nxyz = RM_GetGridCellCount(id)
allocate(species_c(nxyz, nspecies))
status = RM_RunCells(id)
status = RM_GetSpeciesConcentrations(id, species_c)
| integer function phreeqcrm::rm_getspeciescount | ( | integer, intent(in) | id | ) |
The number of aqueous species used in the reaction module. This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | The number of aqueous species, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
nxyz = RM_GetGridCellCount(id)
allocate(species_c(nxyz, nspecies))
status = RM_RunCells(id)
status = RM_GetSpeciesConcentrations(id, species_c)
| integer function phreeqcrm::rm_getspeciesd25 | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out) | diffc | ||
| ) |
Transfers diffusion coefficients at 25C to the array argument (diffc). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. Diffusion coefficients are defined in SOLUTION_SPECIES data blocks, normally in the database file. Databases distributed with the reaction module that have diffusion coefficients defined are phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| diffc | Array to receive the diffusion coefficients at 25 C, m^2/s. Dimension of the array is nspecies, where nspecies is is the number of aqueous species (RM_GetSpeciesCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
allocate(diffc(nspecies))
status = RM_GetSpeciesD25(id, diffc)
| integer function phreeqcrm::rm_getspecieslog10gammas | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | species_log10gammas | ||
| ) |
Transfer log10 aqueous-species activity coefficients to the array argument (species_log10gammas) This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components.
| id | The instance id returned from RM_Create. |
| species_log10gammas | Array to receive the aqueous species concentrations. Dimension of the array is (nxyz, nspecies), where nxyz is the number of user grid cells (RM_GetGridCellCount), and nspecies is the number of aqueous species (RM_GetSpeciesCount). Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
nxyz = RM_GetGridCellCount(id)
allocate(species_log10gammas(nxyz, nspecies))
status = RM_RunCells(id)
status = RM_GetSpeciesLog10Gammas(id, species_log10gammas)
| integer function phreeqcrm::rm_getspecieslog10molalities | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | species_log10molalities | ||
| ) |
Transfer log10 aqueous-species log10 molalities to the array argument (species_log10molalities) To use this method RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components.
| id | The instance id returned from RM_Create. |
| species_log10molalities | Array to receive the aqueous species molalities. Dimension of the array is (nxyz, nspecies), where nxyz is the number of user grid cells (RM_GetGridCellCount), and nspecies is the number of aqueous species (RM_GetSpeciesCount). Values for inactive cells are set to 1e30. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
nxyz = RM_GetGridCellCount(id)
allocate(species_log10molalities(nxyz, nspecies))
status = RM_RunCells(id)
status = RM_GetSpeciesLog10Molalities(id, species_log10molalites)
| integer function phreeqcrm::rm_getspeciesname | ( | integer, intent(in) | id, |
| integer, intent(in) | i, | ||
| character(len=*), intent(out) | name | ||
| ) |
Transfers the name of the ith aqueous species to the character argument (name). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components.
| id | The instance id returned from RM_Create. |
| i | Sequence number of the species in the species list. Fortran, 1 based. |
| name | Character array to receive the species name. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
char*100 name
...
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
do i = 1, nspecies
status = RM_GetSpeciesName(id, i, name)
write(*,*) name
enddo
| integer function phreeqcrm::rm_getspeciessaveon | ( | integer, intent(in) | id | ) |
Returns the value of the species-save property. By default, concentrations of aqueous species are not saved. Setting the species-save property to true allows aqueous species concentrations to be retrieved with RM_GetSpeciesConcentrations, and solution compositions to be set with RM_SpeciesConcentrations2Module.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0, species are not saved; 1, species are saved; negative is failure (See RM_DecodeError). |
save_on = RM_GetSpeciesSaveOn(id)
if (save_on .ne. 0) then
write(*,*) "Reaction module is saving species concentrations"
else
write(*,*) "Reaction module is not saving species concentrations"
end
| integer function phreeqcrm::rm_getspeciesz | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(out) | z | ||
| ) |
Transfers the charge of each aqueous species to the array argument (z). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true.
| id | The instance id returned from RM_Create. |
| z | Array that receives the charge for each aqueous species. Dimension of the array is nspecies, where nspecies is is the number of aqueous species (RM_GetSpeciesCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
allocate(z(nspecies))
status = RM_GetSpeciesZ(id, z)
| integer function phreeqcrm::rm_getstartcell | ( | integer, intent(in) | id, |
| integer, dimension(:), intent(out) | sc | ||
| ) |
Returns an array with the starting cell numbers from the range of cell numbers assigned to each worker.
| id | The instance id returned from RM_Create. |
| sc | Array to receive the starting cell numbers. Dimension of the array is the number of threads (OpenMP) or the number of processes (MPI). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
n = RM_GetThreadCount(id) * RM_GetMpiTasks(id)
allocate(sc(n))
status = RM_GetStartCell(id, sc)
| integer function phreeqcrm::rm_getsurfacename | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves the surface name (such as "Hfo") that corresponds with the surface species name. The lists of surface species names and surface names are the same length. RM_FindComponents must be called before RM_GetSurfaceName. This method may be useful when generating selected output definitions related to surfaces.
| id | The instance id returned from RM_Create. |
| num | The number of the surface name to be retrieved. Fortran, 1 based. |
| name | The surface name associated with surface species num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do i = 1, RM_GetSurfaceSpeciesCount(id)
status = RM_GetSurfaceSpeciesName(id, i, line)
status = RM_GetSurfaceType(id, i, line1)
status = RM_GetSurfaceName(id, i, line2)
input = trim(input) // " " // line // " # " // line1 // line2 // new_line(c)
enddo
| integer function phreeqcrm::rm_getsurfacespeciescount | ( | integer, intent(in) | id | ) |
Returns the number of surface species (such as "Hfo_wOH") in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSurfaceSpeciesCount. This method may be useful when generating selected output definitions related to surfaces.
| id | The instance id returned from RM_Create. |
| The | number of surface species in the initial-phreeqc module. |
do i = 1, RM_GetSurfaceSpeciesCount(id)
status = RM_GetSurfaceSpeciesName(id, i, line)
status = RM_GetSurfaceType(id, i, line1)
status = RM_GetSurfaceName(id, i, line2)
input = trim(input) // " " // line // " # " // line1 // line2 // new_line(c)
enddo
| integer function phreeqcrm::rm_getsurfacespeciesname | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves an item from the surface species list. The list of surface species (for example, "Hfo_wOH") is derived from the list of components (RM_FindComponents) and the list of all surface types (such as "Hfo_w") that are included in SURFACE definitions in the initial-phreeqc module. RM_FindComponents must be called before RM_GetSurfaceSpeciesName. This method may be useful when generating selected output definitions related to surfaces.
| id | The instance id returned from RM_Create. |
| num | The number of the surface type to be retrieved. Fortran, 1 based. |
| name | The surface species name at number num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do i = 1, RM_GetSurfaceSpeciesCount(id)
status = RM_GetSurfaceSpeciesName(id, i, line)
status = RM_GetSurfaceType(id, i, line1)
status = RM_GetSurfaceName(id, i, line2)
input = trim(input) // " " // line // " # " // line1 // line2 // new_line(c)
enddo
| integer function phreeqcrm::rm_getsurfacetype | ( | integer, intent(in) | id, |
| integer, intent(in) | num, | ||
| character(len=*), intent(inout) | name | ||
| ) |
Retrieves the surface site type (such as "Hfo_w") that corresponds with the surface species name. The lists of surface species names and surface species types are the same length. RM_FindComponents must be called before RM_GetSurfaceType. This method may be useful when generating selected output definitions related to surfaces.
| id | The instance id returned from RM_Create. |
| num | The number of the surface type to be retrieved. Fortran, 1 based. |
| name | The surface type associated with surface species num. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do i = 1, RM_GetSurfaceSpeciesCount(id)
status = RM_GetSurfaceSpeciesName(id, i, line)
status = RM_GetSurfaceType(id, i, line1)
status = RM_GetSurfaceName(id, i, line2)
input = trim(input) // " " // line // " # " // line1 // line2 // new_line(c)
enddo
| integer function phreeqcrm::rm_getthreadcount | ( | integer, intent(in) | id | ) |
Returns the number of threads, which is equal to the number of workers used to run in parallel with OPENMP. For the OPENMP version, the number of threads is set implicitly or explicitly with RM_Create. For the MPI version, the number of threads is always one for each process.
| id | The instance id returned from RM_Create. |
| The | number of threads, negative is failure (See RM_DecodeError). |
write(string1, "(A,I)") "Number of threads: ", RM_GetThreadCount(id)
status = RM_OutputMessage(id, string1)
| double precision function phreeqcrm::rm_gettime | ( | integer, intent(in) | id | ) |
Returns the current simulation time in seconds. The reaction module does not change the time value, so the returned value is equal to the default (0.0) or the last time set by RM_SetTime.
| id | The instance id returned from RM_Create. |
| The | current simulation time in seconds. |
write(string, "(A32,F15.1,A)") "Beginning transport calculation ", &
RM_GetTime(id) * RM_GetTimeConversion(id), " days"
status = RM_LogMessage(id, string)
| double precision function phreeqcrm::rm_gettimeconversion | ( | integer, intent(in) | id | ) |
Returns a multiplier to convert time from seconds to another unit, as specified by the user. The reaction module uses seconds as the time unit. The user can set a conversion factor (RM_SetTimeConversion) and retrieve it with RM_GetTimeConversion. The reaction module only uses the conversion factor when printing the long version of cell chemistry (RM_SetPrintChemistryOn), which is rare. Default conversion factor is 1.0.
| id | The instance id returned from RM_Create. |
| Multiplier | to convert seconds to another time unit. |
write(string, "(A32,F15.1,A)") "Beginning transport calculation ", &
RM_GetTime(id) * RM_GetTimeConversion(id), " days"
status = RM_LogMessage(id, string)
| double precision function phreeqcrm::rm_gettimestep | ( | integer, intent(in) | id | ) |
Returns the current simulation time step in seconds. This is the time over which kinetic reactions are integrated in a call to RM_RunCells. The reaction module does not change the time step value, so the returned value is equal to the default (0.0) or the last time step set by RM_SetTimeStep.
| id | The instance id returned from RM_Create. |
| The | current simulation time step in seconds. |
write(string, "(A32,F15.1,A)") " Time step ", &
RM_GetTimeStep(id) * RM_GetTimeConversion(id), " days"
status = RM_LogMessage(id, string)
| integer function phreeqcrm::rm_initialphreeqc2concentrations | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | bc_conc, | ||
| integer, intent(in) | n_boundary, | ||
| integer, dimension(:), intent(in) | bc1, | ||
| integer, dimension(:), intent(in), optional | bc2, | ||
| double precision, dimension(:), intent(in), optional | f1 | ||
| ) |
Fills an array (bc_conc) with concentrations from solutions in the InitialPhreeqc instance. The method is used to obtain concentrations for boundary conditions. If a negative value is used for a cell in bc1, then the highest numbered solution in the InitialPhreeqc instance will be used for that cell. Concentrations may be a mixture of two solutions, bc1 and bc2, with a mixing fraction for bc1 1 of f1 and mixing fraction for bc2 of (1 - f1). A negative value for bc2 implies no mixing, and the associated value for f1 is ignored. If bc2 and f1 are omitted, no mixing is used; concentrations are derived from bc1 only.
| id | The instance id returned from RM_Create. |
| bc_conc | Array of concentrations extracted from the InitialPhreeqc instance. The dimension of bc_conc is (n_boundary, ncomp), where ncomp is the number of components returned from RM_FindComponents or RM_GetComponentCount. |
| n_boundary | The number of boundary condition solutions that need to be filled. |
| bc1 | Array of solution index numbers that refer to solutions in the InitialPhreeqc instance. Size is n_boundary. |
| bc2 | Array of solution index numbers that that refer to solutions in the InitialPhreeqc instance and are defined to mix with bc1. Size is n_boundary. Optional in Fortran. |
| f1 | Fraction of bc1 that mixes with (1-f1) of bc2. Size is (n_boundary). Optional in Fortran. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
nbound = 1
allocate(bc1(nbound), bc2(nbound), f1(nbound))
allocate(bc_conc(nbound, ncomps))
bc1 = 0 ! solution 0 from InitialPhreeqc instance
bc2 = -1 ! no bc2 solution for mixing
f1 = 1.0 ! mixing fraction for bc1
status = RM_InitialPhreeqc2Concentrations(id, bc_conc, nbound, bc1, bc2, f1)

| integer function phreeqcrm::rm_initialphreeqc2module | ( | integer, intent(in) | id, |
| integer, dimension(:,:), intent(in) | ic1, | ||
| integer, dimension(:,:), intent(in), optional | ic2, | ||
| double precision, dimension(:,:), intent(in), optional | f1 | ||
| ) |
Transfer solutions and reactants from the InitialPhreeqc instance to the reaction-module workers, possibly with mixing. In its simplest form, ic1 is used to select initial conditions, including solutions and reactants, for each cell of the model, without mixing. ic1 is dimensioned (nxyz, 7), where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). The dimension of 7 refers to solutions and reactants in the following order: (1) SOLUTIONS, (2) EQUILIBRIUM_PHASES, (3) EXCHANGE, (4) SURFACE, (5) GAS_PHASE, (6) SOLID_SOLUTIONS, and (7) KINETICS. In Fortran, ic1(100, 4) = 2, indicates that cell 99 (0 based) contains the SURFACE definition with user number 2 that has been defined in the InitialPhreeqc instance (either by RM_RunFile or RM_RunString).
It is also possible to mix solutions and reactants to obtain the initial conditions for cells. For mixing, ic2 contains numbers for a second entity that mixes with the entity defined in ic1. F1 contains the mixing fraction for ic1, whereas (1 - f1) is the mixing fraction for ic2. In Fortran, ic1(100, 4) = 2, initial_conditions2(100, 4) = 3, f1(100, 4) = 0.25 indicates that cell 99 (0 based) contains a mixture of 0.25 SURFACE 2 and 0.75 SURFACE 3, where the surface compositions have been defined in the InitialPhreeqc instance. If the user number in ic2 is negative, no mixing occurs. If ic2 and f1 are omitted, no mixing is used, and initial conditions are derived solely from ic1.
| id | The instance id returned from RM_Create. |
| ic1 | Array of solution and reactant index numbers that refer to definitions in the InitialPhreeqc instance. Size is (nxyz,7). The order of definitions is given above. Negative values are ignored, resulting in no definition of that entity for that cell. |
| ic2 | Array of solution and reactant index numbers that refer to definitions in the InitialPhreeqc instance. Nonnegative values of ic2 result in mixing with the entities defined in ic1. Negative values result in no mixing. Size is (nxyz,7). The order of definitions is given above. Optional in Fortran; omitting results in no mixing. |
| f1 | Fraction of ic1 that mixes with (1-f1) of ic2. Size is (nxyz,7). The order of definitions is given above. Optional in Fortran; omitting results in no mixing. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(ic1(nxyz,7), ic2(nxyz,7), f1(nxyz,7))
ic1 = -1
ic2 = -1
f1 = 1.0
do i = 1, nxyz
ic1(i,1) = 1 ! Solution 1
ic1(i,2) = -1 ! Equilibrium phases none
ic1(i,3) = 1 ! Exchange 1
ic1(i,4) = -1 ! Surface none
ic1(i,5) = -1 ! Gas phase none
ic1(i,6) = -1 ! Solid solutions none
ic1(i,7) = -1 ! Kinetics none
enddo
status = RM_InitialPhreeqc2Module(id, ic1, ic2, f1)1))
! No mixing is defined, so the following is equivalent
status = RM_InitialPhreeqc2Module(id, ic1)


| integer function phreeqcrm::rm_initialphreeqc2speciesconcentrations | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(out) | bc_conc, | ||
| integer, intent(in) | n_boundary, | ||
| integer, dimension(:), intent(in) | bc1, | ||
| integer, dimension(:), intent(in), optional | bc2, | ||
| double precision, dimension(:), intent(in), optional | f1 | ||
| ) |
Fills an array (bc_conc) with aqueous species concentrations from solutions in the InitialPhreeqc instance. This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The method is used to obtain aqueous species concentrations for boundary conditions. If a negative value is used for a cell in bc1, then the highest numbered solution in the InitialPhreeqc instance will be used for that cell. Concentrations may be a mixture of two solutions, bc1 and bc2, with a mixing fraction for bc1 of f1 and mixing fraction for bc2 of (1 - f1). A negative value for bc2 implies no mixing, and the associated value for f1 is ignored. If bc2 and f1 are omitted, no mixing is used; concentrations are derived from bc1 only.
| id | The instance id returned from RM_Create. |
| bc_conc | Array of aqueous concentrations extracted from the InitialPhreeqc instance. The dimension of species_c is (n_boundary, nspecies), where nspecies is the number of aqueous species returned from RM_GetSpeciesCount. |
| n_boundary | The number of boundary condition solutions that need to be filled. |
| bc1 | Array of solution index numbers that refer to solutions in the InitialPhreeqc instance. Size is n_boundary. |
| bc2 | Array of solution index numbers that that refer to solutions in the InitialPhreeqc instance and are defined to mix with bc1. Size is n_boundary. Optional in Fortran. |
| f1 | Fraction of bc1 that mixes with (1-f1) of bc2. Size is n_boundary. Optional in Fortran. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
nbound = 1
allocate(bc1(nbound), bc2(nbound), f1(nbound))
allocate(bc_conc(nbound, ncomps))
bc1 = 0 ! solution 0 from InitialPhreeqc instance
bc2 = -1 ! no bc2 solution for mixing
f1 = 1.0 ! mixing fraction for bc1
status = RM_InitialPhreeqc2SpeciesConcentrations(id, bc_conc, nbound, bc1, bc2, f1)

| integer function phreeqcrm::rm_initialphreeqccell2module | ( | integer, intent(in) | id, |
| integer, intent(in) | n_user, | ||
| integer, dimension(:), intent(in) | cell_numbers, | ||
| integer, intent(in) | n_cell | ||
| ) |
A cell numbered n_user in the InitialPhreeqc instance is selected to populate a series of cells. All reactants with the number n_user are transferred along with the solution. If MIX n_user exists, it is used for the definition of the solution. If n_user is negative, n_user is redefined to be the largest solution or MIX number in the InitialPhreeqc instance. All reactants for each cell in the list cell_numbers are removed before the cell definition is copied from the InitialPhreeqc instance to the workers.
| id | The instance id returned from RM_Create. |
| n_user | Cell number refers to a solution or MIX and associated reactants in the InitialPhreeqc instance. A negative number indicates the largest solution or MIX number in the InitialPhreeqc instance will be used. |
| cell_numbers | A list of cell numbers in the user's grid-cell numbering system that will be populated with cell n_user from the InitialPhreeqc instance. |
| n_cell | The number of cell numbers in the cell_numbers list. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate (cell_numbers(2))
cell_numbers(1) = 18
cell_numbers(2) = 19
! n will be the largest SOLUTION number in InitialPhreeqc instance
! copies solution and reactants to cells 18 and 19
status = RM_InitialPhreeqcCell2Module(id, -1, cell_numbers, 2)

| integer function phreeqcrm::rm_loaddatabase | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | db_name | ||
| ) |
Load a database for all IPhreeqc instances–workers, InitialPhreeqc, and Utility. All definitions of the reaction module are cleared (SOLUTION_SPECIES, PHASES, SOLUTIONs, etc.), and the database is read.
| id | The instance id returned from RM_Create. |
| db_name | String containing the database name. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_LoadDatabase(id, "phreeqc.dat")

| integer function phreeqcrm::rm_logmessage | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | str | ||
| ) |
Print a message to the log file.
| id | The instance id returned from RM_Create. |
| str | String to be printed. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
write(string, "(A32,F15.1,A)") "Beginning transport calculation ", &
RM_GetTime(id) * RM_GetTimeConversion(id), " days"
status = RM_LogMessage(id, string)
| integer function phreeqcrm::rm_mpiworker | ( | integer, intent(in) | id | ) |
MPI only. Workers (processes with RM_GetMpiMyself > 0) must call RM_MpiWorker to be able to respond to messages from the root to accept data, perform calculations, and (or) return data. RM_MpiWorker contains a loop that reads a message from root, performs a task, and waits for another message from root. RM_SetConcentrations, RM_RunCells, and RM_GetConcentrations are examples of methods that send a message from root to get the workers to perform a task. The workers will respond to all methods that are designated "workers must be in the loop of RM_MpiWorker" in the MPI section of the method documentation. The workers will continue to respond to messages from root until root calls RM_MpiWorkerBreak.
(Advanced) The list of tasks that the workers perform can be extended by using RM_SetMpiWorkerCallback. It is then possible to use the MPI processes to perform other developer-defined tasks, such as transport calculations, without exiting from the RM_MpiWorker loop. Alternatively, root calls RM_MpiWorkerBreak to allow the workers to continue past a call to RM_MpiWorker. The workers perform developer-defined calculations, and then RM_MpiWorker is called again to respond to requests from root to perform reaction-module tasks.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). RM_MpiWorker returns a value only when RM_MpiWorkerBreak is called by root. |
status = RM_MpiWorker(id)
| integer function phreeqcrm::rm_mpiworkerbreak | ( | integer, intent(in) | id | ) |
MPI only. This method is called by root to force workers (processes with RM_GetMpiMyself > 0) to return from a call to RM_MpiWorker. RM_MpiWorker contains a loop that reads a message from root, performs a task, and waits for another message from root. The workers respond to all methods that are designated "workers must be in the loop of RM_MpiWorker" in the MPI section of the method documentation. The workers will continue to respond to messages from root until root calls RM_MpiWorkerBreak.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_MpiWorkerBreak(id)
| integer function phreeqcrm::rm_openfiles | ( | integer, intent(in) | id | ) |
Opens the output and log files. Files are named prefix.chem.txt and prefix.log.txt based on the prefix defined by RM_SetFilePrefix.
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetFilePrefix(id, "Advect_f90")
status = RM_OpenFiles(id)
| integer function phreeqcrm::rm_outputmessage | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | str | ||
| ) |
Print a message to the output file.
| id | The instance id returned from RM_Create. |
| str | String to be printed. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
write(string1, "(A,I10)") "Number of threads: ", RM_GetThreadCount(id)
status = RM_OutputMessage(id, string1)
write(string1, "(A,I10)") "Number of MPI processes: ", RM_GetMpiTasks(id)
status = RM_OutputMessage(id, string1)
write(string1, "(A,I10)") "MPI task number: ", RM_GetMpiMyself(id)
status = RM_OutputMessage(id, string1)
status = RM_GetFilePrefix(id, string)
write(string1, "(A,A)") "File prefix: ", string
status = RM_OutputMessage(id, trim(string1))
write(string1, "(A,I10)") "Number of grid cells in the user's model: ", RM_GetGridCellCount(id)
status = RM_OutputMessage(id, trim(string1))
write(string1, "(A,I10)") "Number of chemistry cells in the reaction module: ", RM_GetChemistryCellCount(id)
status = RM_OutputMessage(id, trim(string1))
write(string1, "(A,I10)") "Number of components for transport: ", RM_GetComponentCount(id)
status = RM_OutputMessage(id, trim(string1))
| integer function phreeqcrm::rm_runcells | ( | integer, intent(in) | id | ) |
Runs a reaction step for all of the cells in the reaction module. Normally, tranport concentrations are transferred to the reaction cells (RM_SetConcentrations) before reaction calculations are run. The length of time over which kinetic reactions are integrated is set by RM_SetTimeStep. Other properties that may need to be updated as a result of the transport calculations include porosity (RM_SetPorosity), saturation (RM_SetSaturation), temperature (RM_SetTemperature), and pressure (RM_SetPressure).
| id | The instance id returned from RM_Create. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetPorosity(id, por) ! If pore volume changes
status = RM_SetSaturation(id, sat) ! If saturation changes
status = RM_SetTemperature(id, temperature) ! If temperature changes
status = RM_SetPressure(id, pressure) ! If pressure changes
status = RM_SetConcentrations(id, c) ! Transported concentrations
status = RM_SetTimeStep(id, time_step) ! Time step for kinetic reactions
status = RM_RunCells(id)
status = RM_GetConcentrations(id, c) ! Concentrations after reaction
status = RM_GetDensity(id, density) ! Density after reaction
status = RM_GetSolutionVolume(id, volume) ! Solution volume after reaction
| integer function phreeqcrm::rm_runfile | ( | integer, intent(in) | id, |
| integer, intent(in) | workers, | ||
| integer, intent(in) | initial_phreeqc, | ||
| integer, intent(in) | utility, | ||
| character(len=*), intent(in) | chem_name | ||
| ) |
Run a PHREEQC input file. The first three arguments determine which IPhreeqc instances will run the file–the workers, the InitialPhreeqc instance, and (or) the Utility instance. Input files that modify the thermodynamic database should be run by all three sets of instances. Files with SELECTED_OUTPUT definitions that will be used during the time-stepping loop need to be run by the workers. Files that contain initial conditions or boundary conditions should be run by the InitialPhreeqc instance.
| id | The instance id returned from RM_Create. |
| workers | 1, the workers will run the file; 0, the workers will not run the file. |
| initial_phreeqc | 1, the InitialPhreeqc instance will run the file; 0, the InitialPhreeqc will not run the file. |
| utility | 1, the Utility instance will run the file; 0, the Utility instance will not run the file. |
| chem_name | Name of the file to run. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_RunFile(id, 1, 1, 1, "advect.pqi")

| integer function phreeqcrm::rm_runstring | ( | integer, intent(in) | id, |
| integer, intent(in) | workers, | ||
| integer, intent(in) | initial_phreeqc, | ||
| integer, intent(in) | utility, | ||
| character(len=*), intent(in) | input_string | ||
| ) |
Run a PHREEQC input string. The first three arguments determine which IPhreeqc instances will run the string–the workers, the InitialPhreeqc instance, and (or) the Utility instance. Input strings that modify the thermodynamic database should be run by all three sets of instances. Strings with SELECTED_OUTPUT definitions that will be used during the time-stepping loop need to be run by the workers. Strings that contain initial conditions or boundary conditions should be run by the InitialPhreeqc instance.
| id | The instance id returned from RM_Create. |
| workers | 1, the workers will run the string; 0, the workers will not run the string. |
| initial_phreeqc | 1, the InitialPhreeqc instance will run the string; 0, the InitialPhreeqc will not run the string. |
| utility | 1, the Utility instance will run the string; 0, the Utility instance will not run the string. |
| input_string | String containing PHREEQC input. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
string = "DELETE; -all"
status = RM_RunString(id, 1, 0, 1, string) ! workers, initial_phreeqc, utility

| integer function phreeqcrm::rm_screenmessage | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | str | ||
| ) |
Print message to the screen.
| id | The instance id returned from RM_Create. |
| str | String to be printed. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
write(string, "(A32,F15.1,A)") "Beginning reaction calculation ", &
time * RM_GetTimeConversion(id), " days"
status = RM_ScreenMessage(id, string)
| integer function phreeqcrm::rm_setcomponenth2o | ( | integer, intent(in) | id, |
| integer, intent(in) | tf | ||
| ) |
Select whether to include H2O in the component list. The concentrations of H and O must be known accurately (8 to 10 significant digits) for the numerical method of PHREEQC to produce accurate pH and pe values. Because most of the H and O are in the water species, it may be more robust (require less accuracy in transport) to transport the excess H and O (the H and O not in water) and water. The default setting (true) is to include water, excess H, and excess O as components. A setting of false will include total H and total O as components. RM_SetComponentH2O must be called before RM_FindComponents.
| id | The instance id returned from RM_Create. |
| tf | 0, total H and O are included in the component list; 1, excess H, excess O, and water are included in the component list. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetComponentH2O(id, 0)

| integer function phreeqcrm::rm_setconcentrations | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(in) | c | ||
| ) |
Use the vector of concentrations (c) to set the moles of components in each reaction cell. The volume of water in a cell is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and reference volume (RM_SetRepresentativeVolume). The moles of each component are determined by the volume of water and per liter concentrations. If concentration units (RM_SetUnitsSolution) are mass fraction, the density (as specified by RM_SetDensity) is used to convert from mass fraction to per mass per liter.
| id | The instance id returned from RM_Create. |
| c | Array of component concentrations. Size of array is (nxyz, ncomps), where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount), and ncomps is the number of components as determined by RM_FindComponents or RM_GetComponentCount. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(c(nxyz, ncomps))
...
call advect_f90(c, bc_conc, ncomps, nxyz)
status = RM_SetPorosity(id, por) ! If porosity changes
status = RM_SetSaturation(id, sat) ! If saturation changes
status = RM_SetTemperature(id, temperature)) ! If temperature changes
status = RM_SetPressure(id, pressure) ! If pressure changes
status = RM_SetConcentrations(id, c) ! Transported concentrations
status = RM_SetTimeStep(id, time_step) ! Time step for kinetic reactions
status = RM_SetTime(id, time) ! Current time
status = RM_RunCells(id)
status = RM_GetConcentrations(id, c) ! Concentrations after reaction
status = RM_GetDensity(id, density) ! Density after reaction
status = RM_GetSolutionVolume(id, volume) ! Solution volume after reaction


| integer function phreeqcrm::rm_setconcentrations1d | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | c | ||
| ) |
| integer function phreeqcrm::rm_setcurrentselectedoutputusernumber | ( | integer, intent(in) | id, |
| integer, intent(in) | n_user | ||
| ) |
Select the current selected output by user number. The user may define multiple SELECTED_OUTPUT data blocks for the workers. A user number is specified for each data block. The value of the argument n_user selects which of the SELECTED_OUTPUT definitions will be used for selected-output operations.
| id | The instance id returned from RM_Create. |
| n_user | User number of the SELECTED_OUTPUT data block that is to be used. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
do isel = 1, RM_GetSelectedOutputCount(id)
n_user = RM_GetNthSelectedOutputUserNumber(id, isel)
status = RM_SetCurrentSelectedOutputUserNumber(id, n_user)
col = RM_GetSelectedOutputColumnCount(id)
allocate(selected_out(nxyz,col))
status = RM_GetSelectedOutput(id, selected_out)
! Process results here
deallocate(selected_out)
enddo
| integer function phreeqcrm::rm_setdensity | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | density | ||
| ) |
Set the density for each reaction cell. These density values are used when converting from transported mass fraction concentrations (RM_SetUnitsSolution) to produce per liter concentrations during a call to RM_SetConcentrations. They are also used when converting from module concentrations to transport concentrations of mass fraction (RM_GetConcentrations), if RM_UseSolutionDensityVolume is set to false.
| id | The instance id returned from RM_Create. |
| density | Array of densities. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(density(nxyz))
density = 1.0
status = RM_SetDensity(id, density(1))

| integer function phreeqcrm::rm_setdumpfilename | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | dump_name | ||
| ) |
Set the name of the dump file. It is the name used by RM_DumpModule.
| id | The instance id returned from RM_Create. |
| dump_name | Name of dump file. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetDumpFileName(id, "advection_f90.dmp")
dump_on = 1
append = 0
status = RM_DumpModule(id, dump_on, append)
| integer function phreeqcrm::rm_seterrorhandlermode | ( | integer, intent(in) | id, |
| integer, intent(in) | mode | ||
| ) |
Set the action to be taken when the reaction module encounters an error. Options are 0, return to calling program with an error return code (default); 1, throw an exception, in C++, the exception can be caught, for C and Fortran, the program will exit; or 2, attempt to exit gracefully.
| id | The instance id returned from RM_Create. |
| mode | Error handling mode: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
id = RM_create(nxyz, nthreads)
status = RM_SetErrorHandlerMode(id, 2)

| integer function phreeqcrm::rm_seterroron | ( | integer, intent(in) | id, |
| integer, intent(in) | tf | ||
| ) |
Set the property that controls whether error messages are generated and displayed. Messages include PHREEQC "ERROR" messages, and any messages written with RM_ErrorMessage.
| id | The instance id returned from RM_Create. |
| tf | 1, enable error messages; 0, disable error messages. Default is 1. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetErrorOn(rm_id, 1)

| integer function phreeqcrm::rm_setfileprefix | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | prefix | ||
| ) |
Set the prefix for the output (prefix.chem.txt) and log (prefix.log.txt) files. These files are opened by RM_OpenFiles.
| id | The instance id returned from RM_Create. |
| prefix | Prefix used when opening the output and log files. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetFilePrefix(id, "Advect_f90")
status = RM_OpenFiles(id)
| integer function phreeqcrm::rm_setgascompmoles | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(in) | gas_moles | ||
| ) |
Use the array of concentrations (gas_moles) to set the moles of gas components in each reaction cell.
| id | The instance id returned from RM_Create. |
| gas_moles | Array of moles of gas components. Dimensions of the vector are (nxyz, ngas_comps), where ngas_comps is the result of RM_GetGasComponentsCount, and nxyz is the number of user grid cells (RM_GetGridCellCount). If the number of moles is set to a negative number, the gas component will not be defined for the GAS_PHASE of the reaction cell. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
ngas_comps = RM_SetGasComponentsCount(id)
allocate(gas_moles(nxyz, ngas_comps))
...
status = RM_SetGasCompMoles(id, gas_moles)
status = RM_RunCells(id)
| integer function phreeqcrm::rm_setgasphasevolume | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | gas_volume | ||
| ) |
Transfer volumes of gas phases from the array given in the argument list (gas_volume) to each reaction cell. The gas-phase volume affects the pressures calculated for fixed-volume gas phases. If a gas-phase volume is defined with this method for a GAS_PHASE in a cell, the gas phase is forced to be a fixed-volume gas phase.
| id | The instance id returned from RM_Create. |
| gas_volume | Array of gas-phase volumes. Dimension of the array is (nxyz), where nxyz is the number of user grid cells (RM_GetGridCellCount). If an element of the array is set to a negative number, the gas component will not be defined for the GAS_PHASE of the reaction cell. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(gas_volume(nxyz))
...
status = RM_SetGasPhaseVolume(id, gas_volume)
status = RM_RunCells(id)
| integer function phreeqcrm::rm_setmpiworkercallback | ( | integer, intent(in) | id, |
| fcn | |||
| ) |
MPI only. Defines a callback function that allows additional tasks to be done by the workers. The method RM_MpiWorker contains a loop, where the workers receive a message (an integer), run a function corresponding to that integer, and then wait for another message. RM_SetMpiWorkerCallback allows the developer to add another function that responds to additional integer messages by calling developer-defined functions corresponding to those integers. RM_MpiWorker calls the callback function when the message number is not one of the PhreeqcRM message numbers. Messages are unique integer numbers. PhreeqcRM uses integers in a range beginning at 0. It is suggested that developers use message numbers starting at 1000 or higher for their tasks. The callback function calls a developer-defined function specified by the message number and then returns to RM_MpiWorker to wait for another message.
For Fortran, the functions that are called from the callback function can use USE statements to find the data necessary to perform the tasks, and the only argument to the callback function is an integer message argument. RM_SetMpiWorkerCallback must be called by each worker before RM_MpiWorker is called.
The motivation for this method is to allow the workers to perform other tasks, for instance, parallel transport calculations, within the structure of RM_MpiWorker. The callback function can be used to allow the workers to receive data, perform transport calculations, and (or) send results, without leaving the loop of RM_MpiWorker. Alternatively, it is possible for the workers to return from RM_MpiWorker by a call to RM_MpiWorkerBreak by root. The workers could then call subroutines to receive data, calculate transport, and send data, and then resume processing PhreeqcRM messages from root with another call to RM_MpiWorker.
| id | The instance id returned from RM_Create. |
| fcn | A function that returns an integer and has an integer argument. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
Code executed by root:
status = do_something()
Code executed by workers:
status = RM_SetMpiWorkerCallback(id, worker_tasks_f)
status = RM_MpiWorker(id)
Code executed by root and workers:
integer function do_something
implicit none
INCLUDE 'mpif.h'
integer status
integer i, method_number, mpi_myself, mpi_task, mpi_tasks, worker_number;
method_number = 1000
call MPI_Comm_size(MPI_COMM_WORLD, mpi_tasks, status)
call MPI_Comm_rank(MPI_COMM_WORLD, mpi_myself, status)
if (mpi_myself .eq. 0) then
CALL MPI_Bcast(method_number, 1, MPI_INTEGER, 0, MPI_COMM_WORLD, status)
write(*,*) "I am root."
do i = 1, mpi_tasks-1
CALL MPI_Recv(worker_number, 1, MPI_INTEGER, i, 0, MPI_COMM_WORLD, MPI_STATUS_IGNORE, status)
write(*,*) "Recieved data from worker number ", worker_number, "."
enddo
else
CALL MPI_Send(mpi_myself, 1, MPI_INTEGER, 0, 0, MPI_COMM_WORLD, status)
endif
do_something = 0
end function do_something
Code called by workers from method MpiWorker:
integer(kind=C_INT) function worker_tasks_f(method_number) BIND(C, NAME='worker_tasks_f')
USE ISO_C_BINDING
implicit none
interface
integer function do_something
end function do_something
end interface
integer(kind=c_int), intent(in) :: method_number
integer :: status
if (method_number .eq. 1000) then
status = do_something()
endif
worker_tasks_f = 0
end function worker_tasks_f
| integer function phreeqcrm::rm_setpartitionuzsolids | ( | integer, intent(in) | id, |
| integer, intent(in) | tf | ||
| ) |
Sets the property for partitioning solids between the saturated and unsaturated parts of a partially saturated cell.
The option is intended to be used by saturated-only flow codes that allow a variable water table. The value has meaning only when saturations less than 1.0 are encountered. The partially saturated cells may have a small water-to-rock ratio that causes reactions to proceed differently relative to fully saturated cells. By setting RM_SetPartitionUZSolids to true, the amounts of solids and gases are partioned according to the saturation. If a cell has a saturation of 0.5, then the water interacts with only half of the solids and gases; the other half is unreactive until the water table rises. As the saturation in a cell varies, solids and gases are transferred between the saturated and unsaturated (unreactive) reservoirs of the cell. Unsaturated-zone flow and transport codes will probably use the default (false), which assumes all gases and solids are reactive regardless of saturation.
| id | The instance id returned from RM_Create. |
| tf | True, the fraction of solids and gases available for reaction is equal to the saturation; False (default), all solids and gases are reactive regardless of saturation. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetPartitionUZSolids(id, 0)

| integer function phreeqcrm::rm_setporosity | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | por | ||
| ) |
Set the porosity for each reaction cell. The volume of water in a reaction cell is the product of the porosity, the saturation (RM_SetSaturation), and the representative volume (RM_SetRepresentativeVolume).
| id | The instance id returned from RM_Create. |
| por | Array of porosities, unitless. Default is 0.1. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(por(nxyz))
por = 0.2
status = RM_SetPorosity(id, por(1))


| integer function phreeqcrm::rm_setpressure | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | p | ||
| ) |
Set the pressure for each reaction cell. Pressure effects are considered only in three of the databases distributed with PhreeqcRM: phreeqc.dat, Amm.dat, and pitzer.dat.
| id | The instance id returned from RM_Create. |
| p | Array of pressures, in atm. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(pressure(nxyz))
pressure = 2.0
status = RM_SetPressure(id, pressure(1))

| integer function phreeqcrm::rm_setprintchemistrymask | ( | integer, intent(in) | id, |
| integer, dimension(:), intent(in) | cell_mask | ||
| ) |
Enable or disable detailed output for each reaction cell. Printing for a cell will occur only when the printing is enabled with RM_SetPrintChemistryOn and the cell_mask value is 1.
| id | The instance id returned from RM_Create. |
| cell_mask | Array of integers. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). A value of 0 will disable printing detailed output for the cell; a value of 1 will enable printing detailed output for a cell. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(print_chemistry_mask(nxyz))
do i = 1, nxyz/2
print_chemistry_mask(i) = 1
print_chemistry_mask(i+nxyz/2) = 0
enddo
status = RM_SetPrintChemistryMask(id, print_chemistry_mask(1))

| integer function phreeqcrm::rm_setprintchemistryon | ( | integer, intent(in) | id, |
| integer, intent(in) | workers, | ||
| integer, intent(in) | initial_phreeqc, | ||
| integer, intent(in) | utility | ||
| ) |
Setting to enable or disable printing detailed output from reaction calculations to the output file for a set of cells defined by RM_SetPrintChemistryMask. The detailed output prints all of the output typical of a PHREEQC reaction calculation, which includes solution descriptions and the compositions of all other reactants. The output can be several hundred lines per cell, which can lead to a very large output file (prefix.chem.txt, RM_OpenFiles). For the worker instances, the output can be limited to a set of cells (RM_SetPrintChemistryMask) and, in general, the amount of information printed can be limited by use of options in the PRINT data block of PHREEQC (applied by using RM_RunFile or RM_RunString). Printing the detailed output for the workers is generally used only for debugging, and PhreeqcRM will run significantly faster when printing detailed output for the workers is disabled.
| id | The instance id returned from RM_Create. |
| workers | 0, disable detailed printing in the worker instances, 1, enable detailed printing in the worker instances. |
| initial_phreeqc | 0, disable detailed printing in the InitialPhreeqc instance, 1, enable detailed printing in the InitialPhreeqc instances. |
| utility | 0, disable detailed printing in the Utility instance, 1, enable detailed printing in the Utility instance. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetPrintChemistryOn(id, 0, 1, 0) ! workers, initial_phreeqc, utility
| integer function phreeqcrm::rm_setrebalancebycell | ( | integer, intent(in) | id, |
| integer, intent(in) | method | ||
| ) |
Set the load-balancing algorithm. PhreeqcRM attempts to rebalance the load of each thread or process such that each thread or process takes the same amount of time to run its part of a RM_RunCells calculation. Two algorithms are available; one uses individual times for each cell and accounts for cells that were not run because saturation was zero (default), and the other assigns an average time to all cells. The methods are similar, but limited testing indicates the default method performs better.
| id | The instance id returned from RM_Create. |
| method | 0, indicates average times are used in rebalancing; 1 indicates individual cell times are used in rebalancing (default). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetRebalanceByCell(id, 1)

| integer function phreeqcrm::rm_setrebalancefraction | ( | integer, intent(in) | id, |
| double precision, intent(in) | f | ||
| ) |
Sets the fraction of cells that are transferred among threads or processes when rebalancing. PhreeqcRM attempts to rebalance the load of each thread or process such that each thread or process takes the same amount of time to run its part of a RM_RunCells calculation. The rebalancing transfers cell calculations among threads or processes to try to achieve an optimum balance. RM_SetRebalanceFraction adjusts the calculated optimum number of cell transfers by a fraction from 0 to 1.0 to determine the actual number of cell transfers. A value of zero eliminates load rebalancing. A value less than 1.0 is suggested to slow the approach to the optimum cell distribution and avoid possible oscillations when too many cells are transferred at one iteration, requiring reverse transfers at the next iteration. Default is 0.5.
| id | The instance id returned from RM_Create. |
| f | Fraction from 0.0 to 1.0. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetRebalanceFraction(id, 0.5d0)

| integer function phreeqcrm::rm_setrepresentativevolume | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | rv | ||
| ) |
Set the representative volume of each reaction cell. By default the representative volume of each reaction cell is 1 liter. The volume of water in a reaction cell is determined by the procuct of the representative volume, the porosity (RM_SetPorosity), and the saturation (RM_SetSaturation). The numerical method of PHREEQC is more robust if the water volume for a reaction cell is within a couple orders of magnitude of 1.0. Small water volumes caused by small porosities and (or) small saturations (and (or) small representative volumes) may cause non-convergence of the numerical method. In these cases, a larger representative volume may help. Note that increasing the representative volume also increases the number of moles of the reactants in the reaction cell (minerals, surfaces, exchangers, and others), which are defined as moles per representative volume.
| id | The instance id returned from RM_Create. |
| rv | Vector of representative volumes, in liters. Default is 1.0 liter. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
double precision, dimension(:), allocatable :: rv
allocate(rv(nxyz))
rv = 1.0
status = RM_SetRepresentativeVolume(id, rv(1))

| integer function phreeqcrm::rm_setsaturation | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | sat | ||
| ) |
Set the saturation of each reaction cell. Saturation is a fraction ranging from 0 to 1. The volume of water in a cell is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and representative volume (RM_SetRepresentativeVolume). As a result of a reaction calculation, solution properties (density and volume) will change; the databases phreeqc.dat, Amm.dat, and pitzer.dat have the molar volume data to calculate these changes. The methods RM_GetDensity, RM_GetSolutionVolume, and RM_GetSaturation can be used to account for these changes in the succeeding transport calculation. RM_SetRepresentativeVolume should be called before initial conditions are defined for the reaction cells.
| id | The instance id returned from RM_Create. |
| sat | Array of saturations, unitless. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(sat(nxyz))
sat = 1.0
status = RM_SetSaturation(id, sat(1))

| integer function phreeqcrm::rm_setscreenon | ( | integer, intent(in) | id, |
| integer, intent(in) | tf | ||
| ) |
Set the property that controls whether messages are written to the screen. Messages include information about rebalancing during RM_RunCells, and any messages written with RM_ScreenMessage.
| id | The instance id returned from RM_Create. |
| tf | 1, enable screen messages; 0, disable screen messages. Default is 1. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetScreenOn(rm_id, 1)
| integer function phreeqcrm::rm_setselectedoutputon | ( | integer, intent(in) | id, |
| integer, intent(in) | tf | ||
| ) |
Setting determines whether selected-output results are available to be retrieved with RM_GetSelectedOutput. 1 indicates that selected-output results will be accumulated during RM_RunCells and can be retrieved with RM_GetSelectedOutput; 0 indicates that selected-output results will not be accumulated during RM_RunCells.
| id | The instance id returned from RM_Create. |
| tf | 0, disable selected output; 1, enable selected output. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSelectedOutputOn(id, 1) ! enable selected output
| integer function phreeqcrm::rm_setspeciessaveon | ( | integer, intent(in) | id, |
| integer, intent(in) | save_on | ||
| ) |
Sets the value of the species-save property. This method enables use of PhreeqcRM with multicomponent-diffusion transport calculations. By default, concentrations of aqueous species are not saved. Setting the species-save property to 1 allows aqueous species concentrations to be retrieved with RM_GetSpeciesConcentrations, and solution compositions to be set with RM_SpeciesConcentrations2Module. RM_SetSpeciesSaveOn must be called before calls to RM_FindComponents.
| id | The instance id returned from RM_Create. |
| save_on | 0, indicates species concentrations are not saved; 1, indicates species concentrations are saved. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
save_on = RM_SetSpeciesSaveOn(id, 1)

| integer function phreeqcrm::rm_settemperature | ( | integer, intent(in) | id, |
| double precision, dimension(:), intent(in) | t | ||
| ) |
Set the temperature for each reaction cell. If RM_SetTemperature is not called, worker solutions will have temperatures as defined by initial conditions (RM_InitialPhreeqc2Module and RM_InitialPhreeqcCell2Module).
| id | The instance id returned from RM_Create. |
| t | Array of temperatures, in degrees C. Size of array is nxyz, where nxyz is the number of grid cells in the user's model (RM_GetGridCellCount). |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
allocate(temperature(nxyz))
temperature = 20.0
status = RM_SetTemperature(id, temperature(1))

| integer function phreeqcrm::rm_settime | ( | integer, intent(in) | id, |
| double precision, intent(in) | time | ||
| ) |
Set current simulation time for the reaction module.
| id | The instance id returned from RM_Create. |
| time | Current simulation time, in seconds. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetTime(id, time)

| integer function phreeqcrm::rm_settimeconversion | ( | integer, intent(in) | id, |
| double precision, intent(in) | conv_factor | ||
| ) |
Set a factor to convert to user time units. Factor times seconds produces user time units.
| id | The instance id returned from RM_Create. |
| conv_factor | Factor to convert seconds to user time units. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetTimeConversion(id, dble(1.0 / 86400.0)) ! days
| integer function phreeqcrm::rm_settimestep | ( | integer, intent(in) | id, |
| double precision, intent(in) | time_step | ||
| ) |
Set current time step for the reaction module. This is the length of time over which kinetic reactions are integrated.
| id | The instance id returned from RM_Create. |
| time_step | Current time step, in seconds. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetTimeStep(id, time_step)

| integer function phreeqcrm::rm_setunitsexchange | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Sets input units for exchangers. In PHREEQC input, exchangers are defined by moles of exchange sites (Mp). RM_SetUnitsExchange specifies how the number of moles of exchange sites in a reaction cell (Mc) is calculated from the input value (Mp).
Options are 0, Mp is mol/L of RV (default), Mc = Mp*RV, where RV is the representative volume (RM_SetRepresentativeVolume); 1, Mp is mol/L of water in the RV, Mc = Mp*P*RV, where P is porosity (RM_SetPorosity); or 2, Mp is mol/L of rock in the RV, Mc = Mp*(1-P)*RV.
If a single EXCHANGE definition is used for cells with different initial porosity, the three options scale quite differently. For option 0, the number of moles of exchangers will be the same regardless of porosity. For option 1, the number of moles of exchangers will be vary directly with porosity and inversely with rock volume. For option 2, the number of moles of exchangers will vary directly with rock volume and inversely with porosity.
| id | The instance id returned from RM_Create. |
| option | Units option for exchangers: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetUnitsExchange(id, 1)

| integer function phreeqcrm::rm_setunitsgasphase | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Set input units for gas phases. In PHREEQC input, gas phases are defined by moles of component gases (Mp). RM_SetUnitsGasPhase specifies how the number of moles of component gases in a reaction cell (Mc) is calculated from the input value (Mp).
Options are 0, Mp is mol/L of RV (default), Mc = Mp*RV, where RV is the representative volume (RM_SetRepresentativeVolume); 1, Mp is mol/L of water in the RV, Mc = Mp*P*RV, where P is porosity (RM_SetPorosity); or 2, Mp is mol/L of rock in the RV, Mc = Mp*(1-P)*RV.
If a single GAS_PHASE definition is used for cells with different initial porosity, the three options scale quite differently. For option 0, the number of moles of a gas component will be the same regardless of porosity. For option 1, the number of moles of a gas component will be vary directly with porosity and inversely with rock volume. For option 2, the number of moles of a gas component will vary directly with rock volume and inversely with porosity.
| id | The instance id returned from RM_Create. |
| option | Units option for gas phases: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetUnitsGasPhase(id, 1)

| integer function phreeqcrm::rm_setunitskinetics | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Set input units for kinetic reactants.
In PHREEQC input, kinetics are defined by moles of kinetic reactants (Mp). RM_SetUnitsKinetics specifies how the number of moles of kinetic reactants in a reaction cell (Mc) is calculated from the input value (Mp).
Options are 0, Mp is mol/L of RV (default), Mc = Mp*RV, where RV is the representative volume (RM_SetRepresentativeVolume); 1, Mp is mol/L of water in the RV, Mc = Mp*P*RV, where P is porosity (RM_SetPorosity); or 2, Mp is mol/L of rock in the RV, Mc = Mp*(1-P)*RV.
If a single KINETICS definition is used for cells with different initial porosity, the three options scale quite differently. For option 0, the number of moles of kinetic reactants will be the same regardless of porosity. For option 1, the number of moles of kinetic reactants will be vary directly with porosity and inversely with rock volume. For option 2, the number of moles of kinetic reactants will vary directly with rock volume and inversely with porosity.
Note that the volume of water in a cell in the reaction module is equal to the product of porosity (RM_SetPorosity), the saturation (RM_SetSaturation), and representative volume (RM_SetRepresentativeVolume), which is usually less than 1 liter. It is important to write the RATES definitions for homogeneous (aqueous) kinetic reactions to account for the current volume of water, often by calculating the rate of reaction per liter of water and multiplying by the volume of water (Basic function SOLN_VOL).
Rates that depend on surface area of solids, are not dependent on the volume of water. However, it is important to get the correct surface area for the kinetic reaction. To scale the surface area with the number of moles, the specific area (m^2 per mole of reactant) can be defined as a parameter (KINETICS; -parm), which is multiplied by the number of moles of reactant (Basic function M) in RATES to obtain the surface area.
| id | The instance id returned from RM_Create. |
| option | Units option for kinetic reactants: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetUnitsKinetics(id, 1)

| integer function phreeqcrm::rm_setunitsppassemblage | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Set input units for pure phase assemblages (equilibrium phases). In PHREEQC input, equilibrium phases are defined by moles of each phase (Mp). RM_SetUnitsPPassemblage specifies how the number of moles of phases in a reaction cell (Mc) is calculated from the input value (Mp).
Options are 0, Mp is mol/L of RV (default), Mc = Mp*RV, where RV is the representative volume (RM_SetRepresentativeVolume); 1, Mp is mol/L of water in the RV, Mc = Mp*P*RV, where P is porosity (RM_SetPorosity); or 2, Mp is mol/L of rock in the RV, Mc = Mp*(1-P)*RV.
If a single EQUILIBRIUM_PHASES definition is used for cells with different initial porosity, the three options scale quite differently. For option 0, the number of moles of a mineral will be the same regardless of porosity. For option 1, the number of moles of a mineral will be vary directly with porosity and inversely with rock volume. For option 2, the number of moles of a mineral will vary directly with rock volume and inversely with porosity.
| id | The instance id returned from RM_Create. |
| option | Units option for equilibrium phases: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetUnitsPPassemblage(id, 1)

| integer function phreeqcrm::rm_setunitssolution | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Solution concentration units used by the transport model. Options are 1, mg/L; 2 mol/L; or 3, mass fraction, kg/kgs. PHREEQC defines solutions by the number of moles of each element in the solution.
To convert from mg/L to moles of element in the representative volume of a reaction cell, mg/L is converted to mol/L and multiplied by the solution volume, which is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and representative volume (RM_SetRepresentativeVolume). To convert from mol/L to moles of element in the representative volume of a reaction cell, mol/L is multiplied by the solution volume. To convert from mass fraction to moles of element in the representative volume of a reaction cell, kg/kgs is converted to mol/kgs, multiplied by density (RM_SetDensity) and multiplied by the solution volume.
To convert from moles of element in the representative volume of a reaction cell to mg/L, the number of moles of an element is divided by the solution volume resulting in mol/L, and then converted to mg/L. To convert from moles of element in a cell to mol/L, the number of moles of an element is divided by the solution volume resulting in mol/L. To convert from moles of element in a cell to mass fraction, the number of moles of an element is converted to kg and divided by the total mass of the solution. Two options are available for the volume and mass of solution that are used in converting to transport concentrations: (1) the volume and mass of solution are calculated by PHREEQC, or (2) the volume of solution is the product of porosity (RM_SetPorosity), saturation (RM_SetSaturation), and representative volume (RM_SetRepresentativeVolume), and the mass of solution is volume times density as defined by RM_SetDensity. Which option is used is determined by RM_UseSolutionDensityVolume.
| id | The instance id returned from RM_Create. |
| option | Units option for solutions: 1, 2, or 3, default is 1, mg/L. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetUnitsSolution(id, 1)

| integer function phreeqcrm::rm_setunitsssassemblage | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Set input units for solid-solution assemblages. In PHREEQC, solid solutions are defined by moles of each component (Mp). RM_SetUnitsSSassemblage specifies how the number of moles of solid-solution components in a reaction cell (Mc) is calculated from the input value (Mp).
Options are 0, Mp is mol/L of RV (default), Mc = Mp*RV, where RV is the representative volume (RM_SetRepresentativeVolume); 1, Mp is mol/L of water in the RV, Mc = Mp*P*RV, where P is porosity (RM_SetPorosity); or 2, Mp is mol/L of rock in the RV, Mc = Mp*(1-P)*RV.
| id | The instance id returned from RM_Create. |
| option | Units option for solid solutions: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
If a single SOLID_SOLUTION definition is used for cells with different initial porosity, the three options scale quite differently. For option 0, the number of moles of a solid-solution component will be the same regardless of porosity. For option 1, the number of moles of a solid-solution component will be vary directly with porosity and inversely with rock volume. For option 2, the number of moles of a solid-solution component will vary directly with rock volume and inversely with porosity.
status = RM_SetUnitsSSassemblage(id, 1)

| integer function phreeqcrm::rm_setunitssurface | ( | integer, intent(in) | id, |
| integer, intent(in) | option | ||
| ) |
Set input units for surfaces. In PHREEQC input, surfaces are defined by moles of surface sites (Mp). RM_SetUnitsSurface specifies how the number of moles of surface sites in a reaction cell (Mc) is calculated from the input value (Mp).
Options are 0, Mp is mol/L of RV (default), Mc = Mp*RV, where RV is the representative volume (RM_SetRepresentativeVolume); 1, Mp is mol/L of water in the RV, Mc = Mp*P*RV, where P is porosity (RM_SetPorosity); or 2, Mp is mol/L of rock in the RV, Mc = Mp*(1-P)*RV.
If a single SURFACE definition is used for cells with different initial porosity, the three options scale quite differently. For option 0, the number of moles of surface sites will be the same regardless of porosity. For option 1, the number of moles of surface sites will be vary directly with porosity and inversely with rock volume. For option 2, the number of moles of surface sites will vary directly with rock volume and inversely with porosity.
| id | The instance id returned from RM_Create. |
| option | Units option for surfaces: 0, 1, or 2. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetUnitsSurface(id, 1)

| integer function phreeqcrm::rm_speciesconcentrations2module | ( | integer, intent(in) | id, |
| double precision, dimension(:,:), intent(in) | species_conc | ||
| ) |
Set solution concentrations in the reaction cells based on the vector of aqueous species concentrations (species_conc). This method is intended for use with multicomponent-diffusion transport calculations, and RM_SetSpeciesSaveOn must be set to true. The list of aqueous species is determined by RM_FindComponents and includes all aqueous species that can be made from the set of components. The method determines the total concentration of a component by summing the molarities of the individual species times the stoichiometric coefficient of the element in each species. Solution compositions in the reaction cells are updated with these component concentrations.
| id | The instance id returned from RM_Create. |
| species_conc | Array of aqueous species concentrations. Dimension of the array is (nxyz, nspecies), where nxyz is the number of user grid cells (RM_GetGridCellCount), and nspecies is the number of aqueous species (RM_GetSpeciesCount). Concentrations are moles per liter. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_SetSpeciesSaveOn(id, 1)
ncomps = RM_FindComponents(id)
nspecies = RM_GetSpeciesCount(id)
nxyz = RM_GetGridCellCount(id)
allocate(species_c(nxyz, nspecies))
...
status = RM_SpeciesConcentrations2Module(id, species_c(1,1))
status = RM_RunCells(id)
| integer function phreeqcrm::rm_usesolutiondensityvolume | ( | integer, intent(in) | id, |
| integer, intent(in) | tf | ||
| ) |
Determines the volume and density to use when converting from the reaction-module concentrations to transport concentrations (RM_GetConcentrations). Two options are available to convert concentration units: (1) the density and solution volume calculated by PHREEQC are used, or (2) the specified density (RM_SetDensity) and solution volume are defined by the product of saturation (RM_SetSaturation), porosity (RM_SetPorosity), and representative volume (RM_SetRepresentativeVolume). Transport models that consider density-dependent flow will probably use the PHREEQC-calculated density and solution volume (default), whereas transport models that assume constant-density flow will probably use specified values of density and solution volume. Only the following databases distributed with PhreeqcRM have molar volume information needed to accurately calculate density and solution volume: phreeqc.dat, Amm.dat, and pitzer.dat. Density is only used when converting to transport units of mass fraction.
| id | The instance id returned from RM_Create. |
| tf | True indicates that the solution density and volume as calculated by PHREEQC will be used to calculate concentrations. False indicates that the solution density set by RM_SetDensity and the volume determined by the product of RM_SetSaturation, RM_SetPorosity, and RM_SetRepresentativeVolume, will be used to calculate concentrations retrieved by RM_GetConcentrations. |
status = RM_UseSolutionDensityVolume(id, 0)

| integer function phreeqcrm::rm_warningmessage | ( | integer, intent(in) | id, |
| character(len=*), intent(in) | warn_str | ||
| ) |
Print a warning message to the screen and the log file.
| id | The instance id returned from RM_Create. |
| warn_str | String to be printed. |
| IRM_RESULT | 0 is success, negative is failure (See RM_DecodeError). |
status = RM_WarningMessage(id, "Parameter is out of range, using default")
